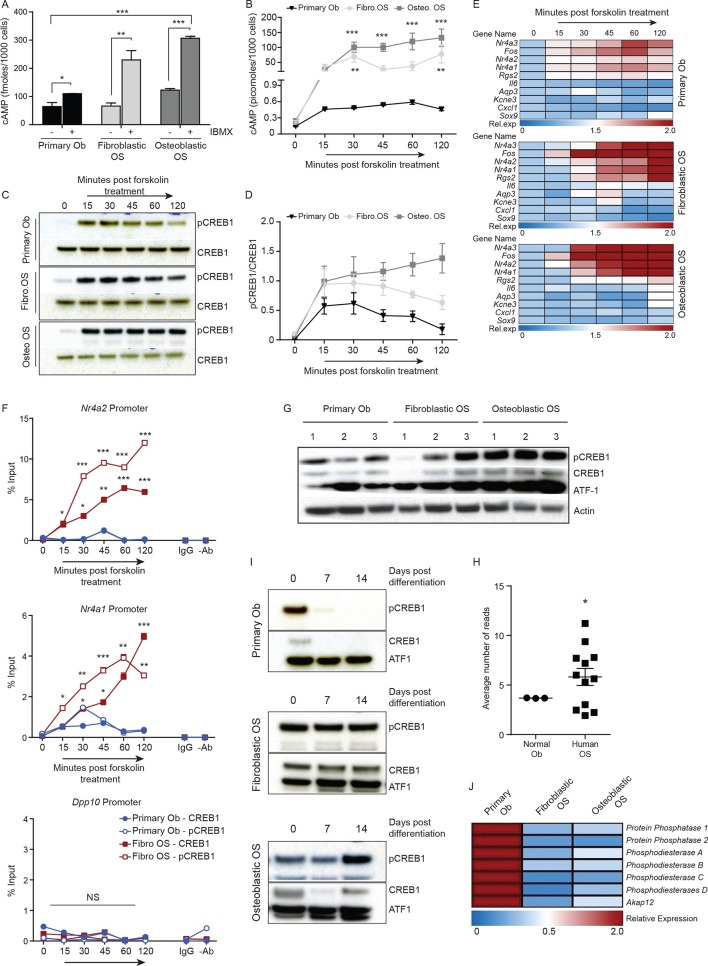
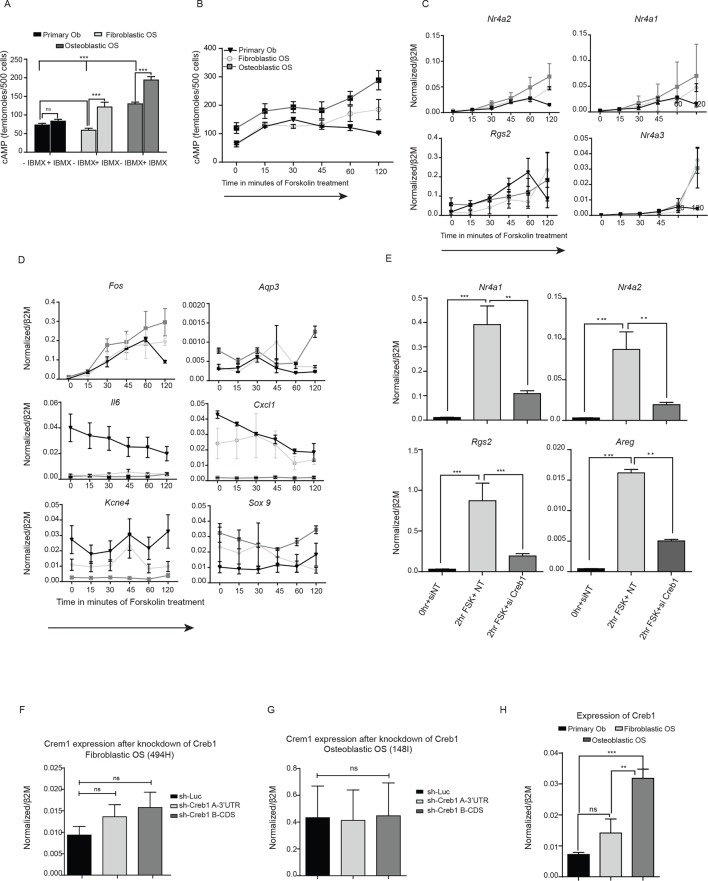
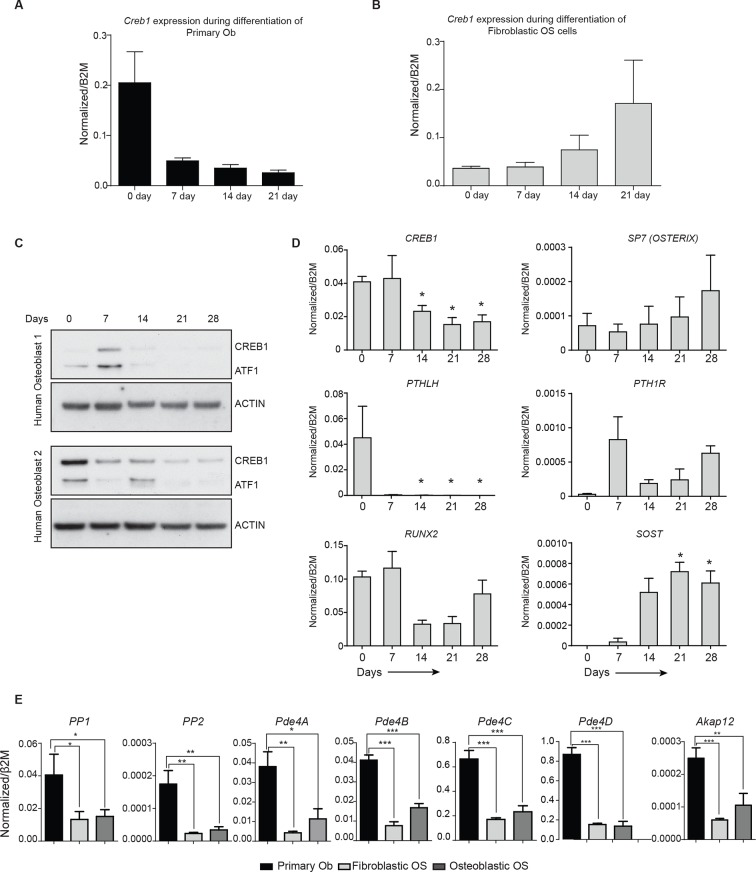
Figure 3. Persistent, elevated cAMP production in OS compared to primary osteoblasts.
(A) cAMP levels in indicated cells (1000 cells per well) with and without IBMX treatment. Data from 3 independent cultures per type, mean ± SEM. (B) Intracellular cAMP levels in indicated cell type following treatment with forskolin; mean ± SEM (n=3 per cell type; 1000 cells per well); statistical significance for OS vs normal Ob; all points of fibroblastic vs osteoblastic OS not significantly different. (C) Western blot and (D) quantification of CREB1/pCREB1 during a time course of cAMP activation by forskolin. Data representative of 2 independent cultures each. (E) Heat map of qPCR data. CREB1 target gene expression in indicated cells; data expressed as relative expression. (F) ChIP analysis of CREB1/pCREB1 on the promoters of indicated genes over a 2 hr time course following stimulation with forskolin. Data is represented as percentage of input. The data from 2 independent cell lines for each subtypes mean occupancy ± SEM (n=2–3 assays per line). (G) Western blot of CREB1/pCREB1 expression in proliferating non stimulated cultures, β-ACTIN used as a loading control. Data representative of 3–4 independent cell lines from each type. (H) CREB1 transcript expression in human osteoblasts and osteosarcoma (data taken from PMID: 25961939). (I) Western CREB1/pCREB1 expression in indicated cell types under differentiative conditions, ATF-1 used as a loading control. (J) Relative expression of negative regulators of cAMP in OS subtypes compared to primary osteoblasts by qPCR and normalized to β2m represented as a heat map (n=3/cell type). *p<0.05, **p<0.001, ***p<0.0001. See Figure 3—figure supplement 1 and Figure 3—figure supplement 2.