Figure 3.
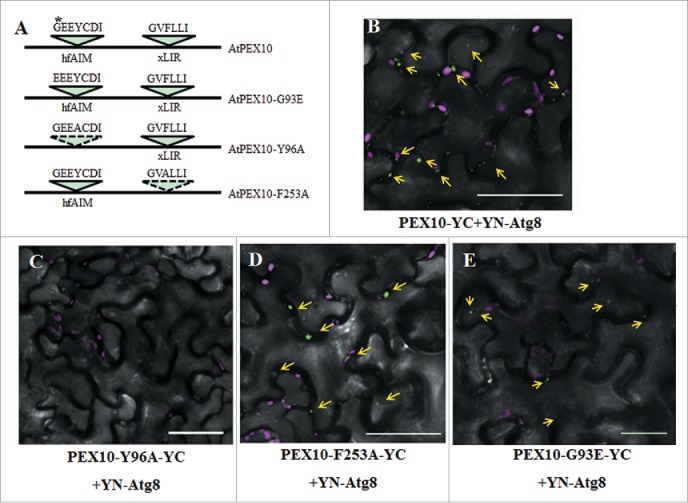
Identification of the functional AIM in AtPEX10. (A) Schematic representation of the AIM sequences predicted by either hfAIM or iLIR in AtPEX10 and their mutants. The referred G-to-E mutation is denoted by a star. (B to E) BiFC analysis was performed following transient coexpression of YN-Atg8f and different variants of PEX10-YC in N. benthamiana leaves. The interaction of wild-type PEX10 with Atg8f results in YFP fluorescence (in green) (B). No interaction was observed when YN-Atg8f was coexpressed with PEX10Y96A-YC (C), demonstrating that the GEEYCDI AIM motif is necessary and sufficient for Atg8f interaction. On the other hand, mutating the AIM motif identified by iLIR did not abolish PEX10 interaction with Atg8f (D), suggesting that this putative AIM is not required for the interaction. The G93E mutation in the hfAIM predicted motif retained the interaction between PEX10 and Atg8f (E). Chloroplast autofluorescence is shown in magenta. Bar: 20 µm.
