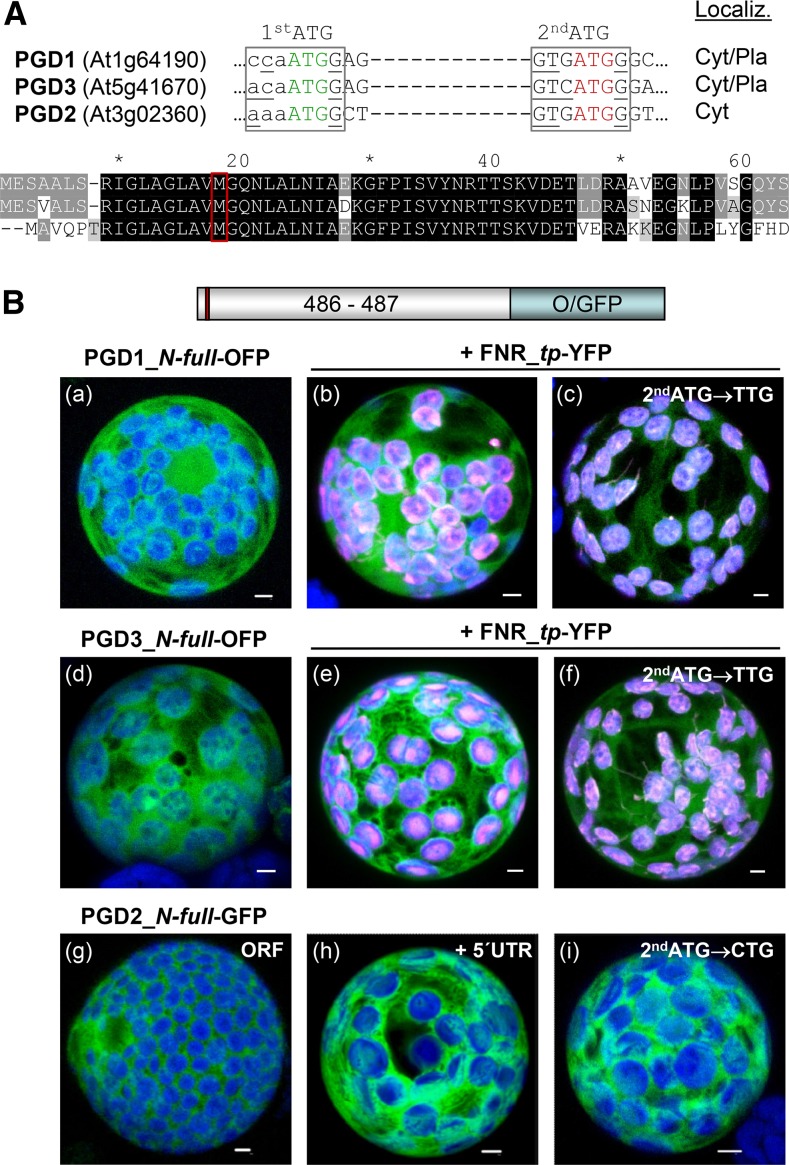
Figure 1.
Localization of full-length PGD1-, PGD3-, and PGD2-reporter fusions. A, Details of the DNA sequence (top) and alignment of the N-terminal ends of the three Arabidopsis PGD isoforms (bottom). A potential second start codon (2ndATG) with Kozak consensus (bases underlined) in all isoforms is highlighted in red. Cyt/Pla, Cytosol/plastids. B, Transient expression of the indicated fusion constructs in tobacco protoplasts (48 h post transfection). PGD3 (a) and PGD1 (d) accumulate in both cytosol and chloroplasts, as confirmed by colocalization with a transit peptide fusion of ferredoxin-NADP reductase (FNR_tp-YFP; b and e). Silent mutagenesis of the second ATG in PGD1 and PGD3 results in enhanced labeling of chloroplasts (c and f). PGD2 labels the cytosol (g) as well as with the 5′ UTR (h) or mutated second ATG (i). Only maximal projections (of approximately 35 single sections) are shown (for single-channel images, see Supplemental Figs. S2 and S4). Candidate fusions are in green, chloroplast marker (FNR) is in magenta, and chlorophyll autofluorescence is in blue. Colocalization and very close signals (less than 200 nm) appear white in merged images. ORF, Open reading frame. Bars = 3 µm.