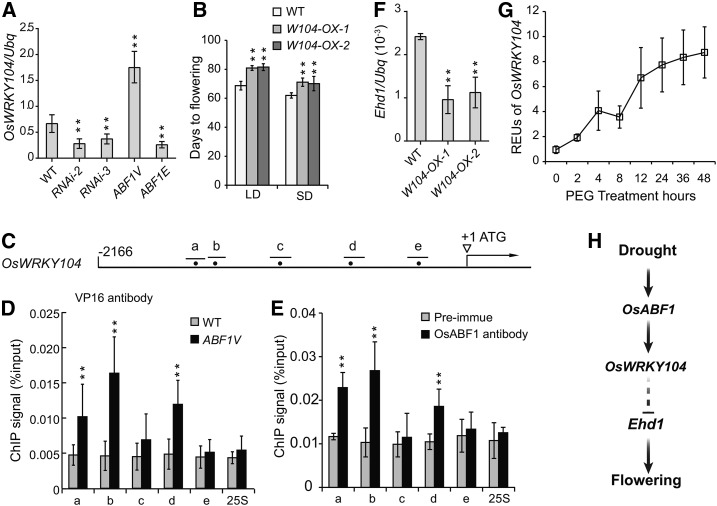
Figure 7.
OsWRKW104 is a direct target of OsABF1that delays flowering. A, qRT-PCR analysis of OsWRKW104 expression in indicated lines. The means ± sd (Student’s t tests, **P < 0.01, n = 3) are shown. B, Flowering days of OsWRKW104 overexpression lines. The means ± sd (n = 15) are shown. C, A diagram representing the promoter region of OsWRKW104 gene. The bars represent the distribution of DNA fragments containing ACGT core as indicated by dots. D, Verification of ABF1V direct binding sites in the OsWRKW104 promoter by ChIP-qPCR analysis. ChIP samples were prepared using ABF1V and wild-type plants, precipitated with anti-VP16 antibody, and subjected to qPCR analysis. Results of ChIP-qPCR were quantified by normalization of the immunoprecipitation signal with the corresponding input signal. The binding to 25S rDNA was used as negative control. The means ± sd (n = 3) are shown. E, Verification of OsABF1 binding sites in the OsWRKW104 promoter by ChIP-qPCR analysis. ChIP samples were prepared using wild-type rice and precipitated with anti-OsABF1 antibody or with the preimmune serum as negative control. The means ± sd (n = 3) are shown. F, qRT-PCR analysis of Ehd1 expression in OsWRKW104 overexpression lines. Values were shown as mean ± sd (n = 3). G, Dynamic transcription of OsWRKW104 in wild type under PEG treatment for the indicated periods. The seedlings were grown in continuous light for 3 weeks and then subjected to PEG treatment. The REUs were calculated by the formula: [(OsWRKW104/ubq) of each time point]/ [(OsWRKW104/ubq) of time point 0]. H, A working model depicts how OsABF1 modulates flowering time in response to drought stress. The transcription of OsABF1 is up-regulated under water deficit. Consequently, OsABF1 instigates the expression of OsWRKW104, which further delays flowering through inhibiting Ehd1 expression.