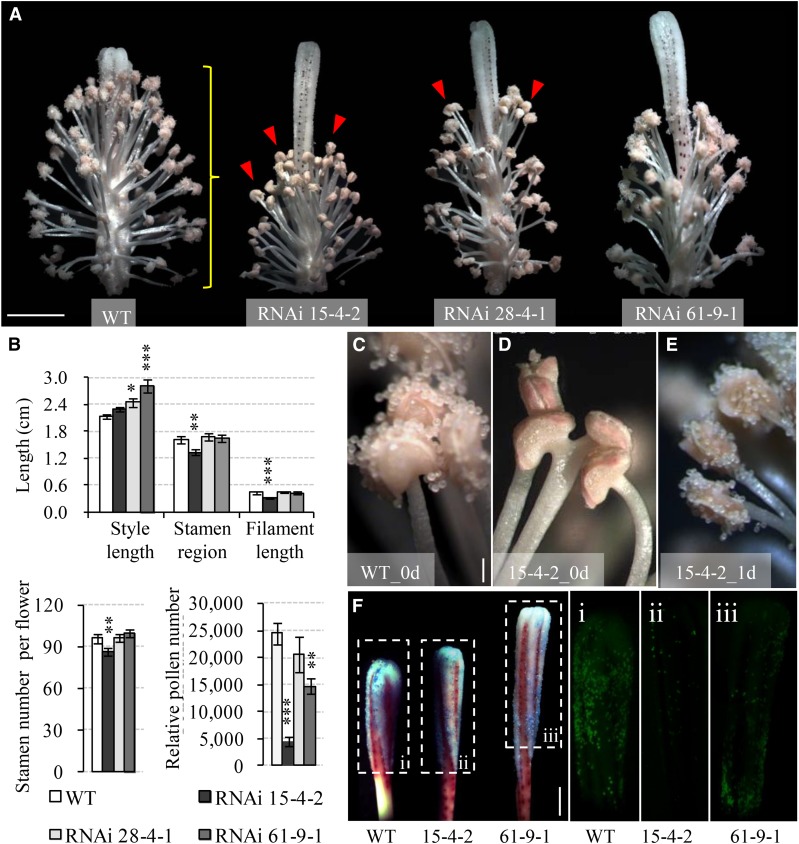
Figure 3.
Silencing GhVIN1 in cotton disrupted style and stamen development, delayed anther dehiscence, and reduced pollen number. A, Representative images of 0-d transgenic flowers with petals removed, showing the uncoordinated style protrusion and stamen development in the RNAi plants compared with the wild type (WT). The red arrowheads indicate indehiscent anthers. The yellow brace indicates the stamen region measured in B. B, GhVIN1-RNAi flowers displayed longer styles or shorter filaments, decreased stamen region, and lower stamen and pollen numbers per flower. Each value is the mean ± se, with data collected from eight flowers of four individual plants for each line. Asterisks denote significant differences (one-way ANOVA; *, P < 0.05; **, P < 0.01; and ***, P < 0.001) between RNAi and wild-type plants. C to E, Anther dehiscence occurred on the day of flowering in the wild type (C) but not in RNAi line 15-4-2 (D). The latter dehisced 1 d later (E). F, Fewer pollens were detected in transgenic styles compared with the wild type on the day of flowering. Styles were stained with Aniline Blue (left; bright field) and viewed under UV light to show the fluorescence emitted from the stained pollen grains in the boxed regions (i–iii). Bars = 1 cm in A, 200 μm in C, and 5 mm in F. The scales in D and E are the same as that in C.