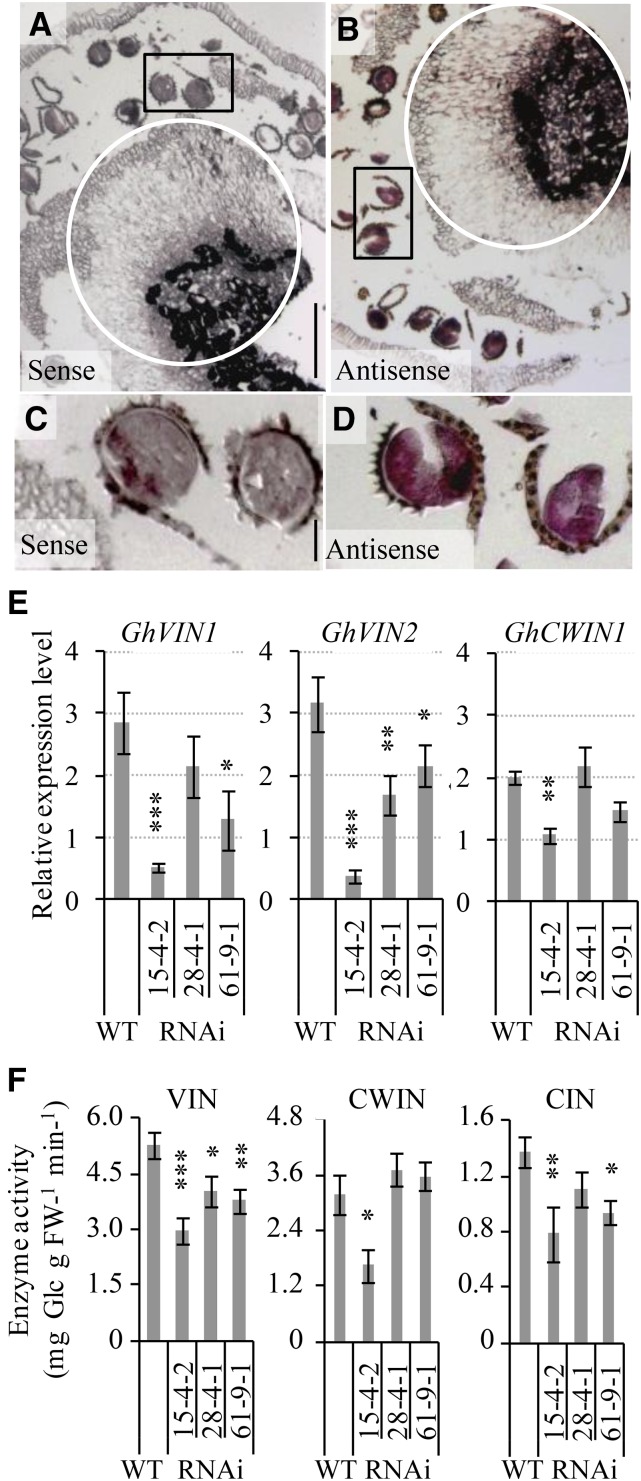
Figure 6.
GhVIN transcripts were abundant at the anther-filament joint region and in pollens of wild-type cotton stamen but were evidently reduced in GhVIN1-RNAi lines, resulting in decreased VIN activities. A to D, A longitudinal section of −1-d wild-type cotton anther hybridized with a sense (A) or an antisense (B) RNA probe for GhVINs. Note the GhVIN mRNA signals in the circled anther-filament joint area in B compared with the same region in the sense control in A. C and D are magnified views of pollens in the boxed areas of A and B, respectively, showing strong GhVIN mRNA signals in wild-type pollens. E, qPCR analyses of GhVIN1, GhVIN2, and GhCWIN1 transcripts in −1-d wild-type (WT) and transgenic stamen. Data represent means ± se (n ≥ 6). F, VIN, CWIN, and CIN activities in −1-d wild-type and RNAi stamen. Each value is the mean ± se (n = 4). Asterisks in E and F indicate significant differences (one-way ANOVA; *, P < 0.05; **, P < 0.01; and ***, P < 0.001) between RNAi and wild-type plants. FW, Fresh weight.