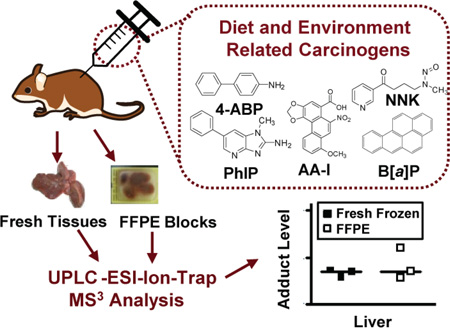
Abstract
DNA adducts are a measure of internal exposure to genotoxicants and an important biomarker for human risk assessment. However, the employment of DNA adducts as biomarkers in human studies is often restricted because fresh frozen tissues are not available. In contrast, formalin-fixed paraffin embedded (FFPE) tissues with clinical diagnosis are readily accessible. Recently, our laboratory reported that DNA adducts of aristolochic acid, a carcinogenic component of Aristolochia herbs used in traditional Chinese medicines world-wide, can be recovered quantitatively from FFPE tissues. In this study, we have evaluated the efficacy of our method for retrieval of DNA adducts from archived tissue by measuring DNA adducts derived from four other classes of human carcinogens: polycyclic aromatic hydrocarbons (PAHs), aromatic amines, heterocyclic aromatic amines (HAAs), and N-nitroso compounds (NOCs). The deoxyguanosine (dG) adducts of the PAH benzo[a]pyrene (B[a]P), 10-(deoxyguanosin-N2-yl)-7,8,9-trihydroxy-7,8,9,10-tetrahydro-benzo[a]pyrene (dG-N2-B[a]PDE); the aromatic amine 4-aminobiphenyl (4-ABP), N-(deoxyguanosin-8-yl)-4-ABP (dG-C8-4-ABP); the HAA 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), N-(deoxyguanosin-8-yl)-PhIP (dG-C8-PhIP); and the dG adducts of the NOC, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), O6-methyl-dG (O6-Me-dG) and O6-pyridyloxobutyl-dG (O6-POB-dG) formed in liver, lung, bladder, pancreas, or colon were recovered in comparable yields from fresh frozen and FFPE preserved tissues of rodents treated with the procarcinogens. Quantification was achieved by Ultra-Performance Liquid Chromatography-Electrospray Ionization-Ion Trap-Multistage Mass Spectrometry (UPLC-ESI-IT-MS3). These advancements in the technology of DNA adduct retrieval from FFPE tissue clear the way for the use of archived pathology samples in molecular epidemiology studies designed to assess the causal role of exposures to hazardous chemicals with cancer risk.
TOC
INTRODUCTION
Humans are continuously exposed to genotoxicants in the environment and diet, and to electrophiles produced endogenously.1,2 Covalent modification of DNA by chemicals results in mutations or other genetic changes and can initiate chemical carcinogenesis.3 DNA adducts are employed for interspecies extrapolation of toxicity data of chemicals and for human risk assessment. The identification and quantitation of DNA adducts often is the first step in elucidating the potential role of a genotoxic chemical in the etiology of human cancer.4,5 Mass Spectrometry (MS) stands out from other methods of DNA adduct analysis for its power in quantifying adducts at parts-per-trillion level, obtaining adduct structural information at the same time.6–8 However, the employment of DNA adduct biomarkers in large-scale population studies has been impeded by the paucity of fresh frozen biospecimens. Conversely, formalin-fixed paraffin-embedded (FFPE) tissues are routinely archived and often accessible for biomarker research.
Formalin fixation is the standard technique for preservation of tissues and has been used world-wide for this purpose for over a century.9 Histopathological and immunohistochemical characterization of FFPE specimens have been reported for almost every disease.10 DNA recovered from FFPE tissues is used widely for molecular genetic studies;11 high-quality DNA extracted from FFPE tissues can serve as a template for real-time Reverse-Transcriptase Polymerase Chain Reaction.12 Previously, our laboratory demonstrated that DNA adducts of aristolochic acid (AA), a potent human carcinogen present in Aristolochia herbs used in traditional Chinese medicines13 can be retrieved quantitatively from FFPE tissues, then measured by an Ultra-Performance Liquid Chromatography coupled-Electrospray Ionization-Ion Trap-Multistage Mass Spectrometry (UPLC-ESI-IT-MS3).14,15 AA found in Aristolochia sp. are responsible for the clinical syndromes known formerly as Balkan endemic nephropathy (BEN)13,16 and Chinese herbs nephropathy.17 Both diseases are associated with a high incidence of urothelial carcinomas of the upper urinary tract,13,16 constituting a disease entity now termed aristolochic acid nephropathy (AAN).18 The major DNA adduct of AA, 7-(deoxyadenosin-N6-yl)-aristolactam (dA-AL-I) is responsible for the characteristic A-T transversion in the TP53 tumor suppressor gene and mutations throughout the tumor genome in patients with AA-induced urothelial carcinomas of the upper urinary tract.13,19 We have successfully employed a commercially available silica-based kit used for cancer genomics to isolate DNA from FFPE tissue for mass spectrometric measurements of dA-AL-I.15
In this study, we sought to determine whether our method for DNA retrieval could also be used to quantify DNA adducts formed by other environmental and dietary carcinogens. Four classes of ubiquitous carcinogens were selected for study: polycyclic aromatic hydrocarbons (PAHs), aromatic amines, heterocyclic aromatic amines (HAAs), and N-nitroso compounds (NOCs) These chemicals occur in the environment and in tobacco smoke and are formed during the cooking of meat.20,21,22 PAH are formed from the incomplete combustion of organic matter, which occurs during the cooking of meat with charcoal, or as ubiquitous environmental pollutants.20 Benzo[a]pyrene (B[a]P) is a prototypical PAH and classified as a Group 1 human carcinogen by the International Agency for Research on Cancer (IARC).23 The biologically active metabolite of B[a]P, 7β,8α-dihydroxy-9α,10α,-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene (B[a]PDE), reacts with DNA at the N2 position of dG, to form 10-(deoxyguanosin-N2-yl)-7,8,9-trihydroxy-7,8,9,10-tetrahydrobenzo[a]pyrene (dG-N2-B[a]PDE).24,25 This adduct induces a G→T transversion mutation at codon 273 of the tumor suppressor gene TP53 and contributes to the initiation of lung cancer in smokers.26 Aromatic amines and HAAs are structurally related classes of carcinogens. Historically, some aromatic amines occurred as contaminants in industrial processes such as dye, rubber and fertilizer, but today the most common and widely spread source of exposure of aromatic amines occurs through tobacco smoke.21 4-Aminobiphenyl (4-ABP) is a human urinary bladder carcinogen that arises in tobacco smoke21 and also occurs as a contaminant of hair dyes27 and in the atmosphere.28 2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) is the most-mass abundant carcinogenic HAA formed during the high-temperature cooking of meat.22 Levels of PhIP can vary from several parts-per-billion (ppb) up to 500 ppb.22,29 PhIP, together with other HAAs formed during the cooking of meats, have been linked to several dietary-associated cancers at sites such as colon, prostate, pancreas, and mammary gland.30 The primary DNA adducts of 4-ABP and PhIP are formed at the C8 position of the purine base of dG, producing N-(2′-deoxyguanosin-8-yl)-4-aminobiphenyl (dG-C8-4-ABP) and N-(2′-deoxyguanosin-8-yl)-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (dG-C8-PhIP).31,32 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), listed as Group 1 human carcinogen by IARC, is a tobacco-specific NOC and a potent lung carcinogen.33 NNK undergoes metabolism by cytochrome P450 to form reactive intermediates that form methyl, pyridyloxobutyl (POB), and pyridylhydroxybutyl adducts with dG and other deoxynucleosides.34 O6-Methyl-2′-deoxyguanosine (O6-Me-dG) and O6-[4-(3-pyridyl)-4-oxobut-1-yl]-2′-deoxyguanosine (O6-POB-dG) contribute to the carcinogenic properties of NNK.35,36 The G→A transition mutation in codon 12 of Kras, induced by O6-MedG, plays an important role in mouse lung carcinogenesis, whereas both DNA methylation and pyridyloxobutylation, by NNK, are important in rat lung tumorigenesis.37
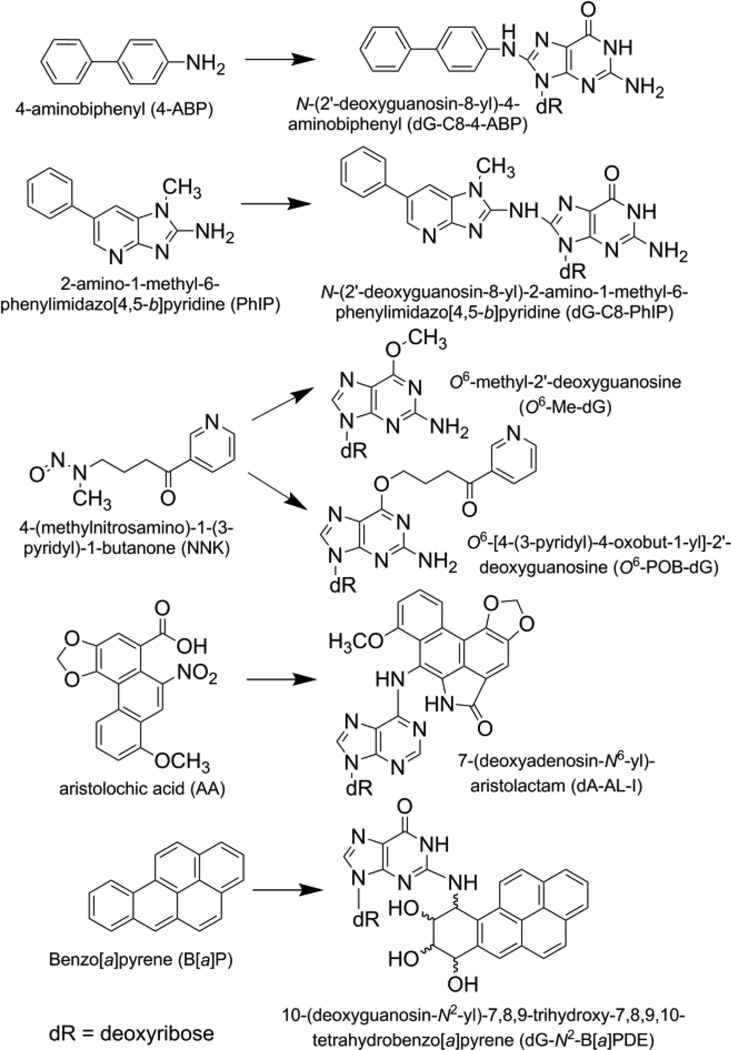
The structures, full names and abbreviations of the carcinogens and their targeted DNA adducts investigated in this study are depicted in Figure 1. The DNA adducts of these carcinogens are well characterized, and adduct levels in rodents have been reported in the literature. 24,25,31,32,34–36 Moreover, the DNA adducts formed from these carcinogens encompass a wide range of chemical properties, including polarity, hydrophobicity, and different ionization efficiencies under the electrospray ionization process employed for liquid chromatography/mass spectrometry (LC/MS). The diversity in chemical structures and properties of DNA adducts reflect the great analytical challenges in DNA adduct analysis.
Figure 1.
Structures, names and abbreviations of the carcinogens and their adducts investigated in this study.
EXPERIMENTAL SECTION
Caution: AA-I, 4-ABP, B[a]P and NNK are human carcinogens; and PhIP is a potential human carcinogen. All of these chemicals should be handled with caution in a well-ventilated fume hood with the appropriate protective clothing.
Materials
PhIP was purchased from Toronto Research Chemicals (Toronto, Canada). 4-ABP, B[a]P, ethanol (200 proof), p-xylene, calf thymus DNA (CT DNA), DNase I (Type IV, bovine pancreas), alkaline phosphatase (Escherichia coli), nuclease P1 (from Penicillium citrinum), RNase A (bovine pancreas) and RNase T1 (Aspergillus oryzae) were purchased from Sigma-Aldrich (St. Louis, MO). Phosphodiesterase I (Crotalus adamanteus venom) and adenosine deaminase were purchased from Worthington Biochemicals Corp. (Newark, NJ). Ten-percent neutral-buffered formalin (NBF), Optima™ LC-MS grade HCO2H, CH3CN, CH3OH and H2O, were purchased from Fisher Chemical (Pittsburgh, PA). 8-Methoxy-6-nitrophenanthro[3,4-d]1,3-dioxole-5-carboxylic acid (AA-I) was a gift from Dr. Horacio Priestap, Department of Biological Sciences, Florida International University. NNK, O6-[4-oxo-4-(3-pyridyl)-butyl]-2′-deoxyguanosine (O6-POB-dG) and [pyridine-2H4]-O6-pyridyloxobutyl-2′-deoxyguanosine ([Pyridine-2H4]-O6-POB-dG),38,39 unlabeled and labelled [13C10]-dG-C8-PhIP, [13C10]-dG-N2-B[a]PDE, and [13C10]-dG-C8-4-ABP,24,32,40 7-(2′-Deoxyadenosin-N6-yl)-aristolactam (dA-AL-I) and [15N5]-dA-AL-I,14 and O6-Methyl-2′-deoxyguanosine (O6-Me-dG) and [2H3C]-O6-Me-dG were synthesized as described.41 ZR FFPE DNA MiniPrep™ kit was purchased from Zymo Research (Irvine, CA). Microliter CapLC vials with silanized inserts were purchased from Wheaton (Millville, NJ). Strata-X solid-phase extraction (SPE) cartridge (33 µm Polymeric Reversed Phase, 10 mg/1 mL) was purchased from Phenomenex (Torrance, CA).
Animal Dosing
The guidelines established by the National Institutes of Health Office of Laboratory Animal Welfare were adhered to for the use of animals with housing conditions described previously.14,15 All protocols were reviewed and approved by the University of Minnesota Institutional Animal Care and Use Committee. The protocol for animal dosing of AA-I has been described.15 The animal strains and dosing levels of carcinogens are summarized in Table S1 and briefly reported here. Male Wistar outbred rats (Harlan Labs, Indianapolis, IN) were dosed by intraperitoneal (i.p.) injection with PhIP (10 mg/kg body weight (BW) in 0.3 mL DMSO). Female A/J mice (Jackson Laboratory, Bar Harbor, ME), were dosed two times at a 24 h interval, by i.p. injection with NNK (80 mg/kg BW in 0.1 mL 0.9% saline). Male B6C3F1 mice (Jackson Laboratory, Bar Harbor, ME) were dosed by i.p. injection with 4-ABP (40 mg/kg BW in 0.1 mL 80% DMSO). Male B6C3F1 mice (Jackson Laboratory, Bar Harbor, ME) were dosed through i.p. injection with B[a]P (100 mg/kg BW in 0.1 mL DMSO). Animals were euthanized by asphyxiation with CO2 after 24 h except for dose treatment with NNK, where animals were sacrificed 3 h after the second dose. The liver, pancreas, lung and bladder were rinsed with chilled PBS, snap frozen on dry ice and stored at −80 °C. The colon was slit open, and rinsed with ice-cold PBS to remove fecal debris, before the mucosal layer was isolated as previously described.42 For formalin fixation, the liver, lung, and pancreas were rinsed with chilled PBS, cut into 0.6 mm thickness pieces, wiped dry with Kimwipes and incubated in 10% NBF for 24 or 48 h at room temperature. The ratio of fixative to tissue or colon mucosal layer was at least 10:1 (w:w). Thereafter, tissues underwent serial dehydration with ethanol followed by p-xylene wash, and then embedded in paraffin using a Sakura Tissue Tek VIP5 tissue processor (Torrance, CA).
DNA Retrieval from Fresh Frozen Tissues
The tissues were thawed on ice and homogenized in TE buffer (50 mM Tris-HCl buffer, pH 8.0, and 10 mM EDTA) containing 10 mM β-mercaptoethanol (βME) with a Wheaton Potter-Elvehjem tissue homogenizer (Millville, NJ). The homogenized tissues (equivalent of 40 mg of wet tissue weight) were centrifuged at 3000 x g for 10 min at 4 °C. DNA was isolated from the pellet by phenol/chloroform extraction method, following digestion with RNase and proteinase K digestion at 37 °C for 2 h.15 Alternatively, the proteinase K digestion was incubated overnight at 50 °C , employing the conditions described for the FFPE DNA Miniprep™ kit (see below).
DNA Retrieval from FFPE Tissues
The procedure used here has been reported previously.15 In brief, the tissues were removed from the paraffin block with a scalpel, and submerged in p-xylene to remove the residual paraffin. The tissues were rehydrated and homogenized in TE buffer containing 10 mM βME and the equivalent of 25 mg dry weight of tissue was processed with the ZR FFPE DNA MiniPrep™ Kit, following the manufacturer’s instructions with minor modifications. An overnight digestion of tissue with proteinase K (0.2 mg) was performed at 50 °C to ensure complete reversal of the cross-linked DNA.
Enzymatic Digestion of DNA
The amounts of DNA and the levels of internal standards (IS) spiked into the DNA prior to digestion are summarized in Table S1. DNA samples were incubated at 37 °C with DNase I and nuclease P1 for 3.5 h, followed by digestion with phosphodiesterase I and alkaline phosphatase overnight. The samples were concentrated to dryness by vacuum centrifugation and dissolved in 50% DMSO for UPLC-ESI-IT-MS3 analysis. The injection volumes and resuspension volumes are summarized in Table S1. The DNA samples from NNK-treated mice were further processed as reported below.
dA-AL-I, dG-N2-B[a]PDE, dG-C8-4-ABP and dG-C8-PhIP Measurement by UPLC-ESI-IT-MS3
The analyses were conducted with a nanoACQUITY UPLC system (Waters Corporation, Milford, MA) interfaced with an Advance CaptiveSpray source (Michrom Bioresource Inc., Auburn, CA), and the Velos Pro Ion-Trap mass spectrometer (Thermo Fisher, San Jose, CA). A Waters UPLC 2G-V/V Symmetry C18 trap column (180 µm x 20 mm, 5 µm) was used for online sample enrichment of DNA adducts. A Magic C18 AQ column (0.1 mm x 150 mm, 3 µm, 100 Å, Michrom Bioresources Inc.) was employed for chromatography. The LC solvents were: (A) 0.01% HCO2H and (B) 0.01% HCO2H and 95% CH3CN. Adduct measurements were performed in the positive ion mode at the MS3 scan stage. The DNA digests were injected onto the trap column and washed with solvent A for 3 min at a flow rate of 12 µL/min. The amount of DNA digest loaded on-column is summarized in Table S1. After online enrichment, adducts were back-flushed onto the Magic C18 AQ column at a flow rate of 1 µL/min. A linear gradient of 1% to 99% B over 9 min was applied for the separation, followed by 4 min washing at 99% B.
Xcalibur version 2.1 was used for data acquisition and analysis. The MS parameters were optimized as previously described.43 Adducts were quantified at the MS3 scan stage with Extracted Ion Chromatograms (EIC) of the two most abundant fragment ions. The MS3 transitions employed for adduct quantification were m/z 543.2 → 427.2 → 292.1, 293.1 and 412.1 (dA-AL-I); 548.2 → 432.2 → 292.1, 293.1 and 417.1 ([15N5]-dA-AL-I); 435.2 → 319.1 → 277.1 and 302.1 (dG-C8-4-ABP), 445.2 → 324.1 → 281.1 and 307.1 ([13C10]-dG-C8-4-ABP); 490.2 → 374.1 → 329.1 and 357.1 (dG-C8-PhIP), 500.2 → 379.1 → 333.1 and 362.1 ([13C10]-dG-C8-PhIP); 570.2 → 454.1 → 285.1 and 303.1 (dG-N2-B[a]PDE) and 580.2 → 459.1 → 285.1 and 303.1 ([13C10]-dG-N2-B[a]PDE).
O6-Me-dG and O6-POB-dG Enrichment by Solid-Phase Extraction (SPE) and Quantitation by UPLC-ESI-IT-MS3
The DNA digest of tissues of mice treated with NNK was further incubated with 0.5 mU adenosine deaminase for 1 h at room temperature to convert dA into deoxyinosine (dI).44 O6-Me-dG and O6-POB-dG were enriched by a Strata-X SPE cartridge pre-conditioned with CH3OH and H2O before sample loading. The DNA digest was washed with H2O (2 mL), 10% CH3OH (1 mL), 25% CH3OH (2× 1 mL), and adducts were eluted with CH3OH (1 mL). The eluents were dried by vacuum centrifugation, resuspended in CH3OH (100 µL), transferred to CapLC vials and dried by vacuum centrifuge again. Water was added to reconstitute the sample for LC-MS analysis (20 µL). The UPLC-ESI/MS system described above was used in the direct injection mode for the quantification of O6-Me-dG and O6-POB-dG. The injection volume was 5 µL. The MS3 transitions employed were m/z 282.1 → 166.1 → 149.1 (O6-Me-dG) and 285.1 → 169.1 → 152.1 ([2H3]-O6-Me-dG); 415.2 → 299.1 → 148.1 and 152.1 (O6-POB-dG) and 419.2 → 303.1 → 152.1 ([pyridine-2H4]-O6-POB-dG).
Calibration Curves
The calibration curves were constructed at five levels. Isotope labeled and unlabeled dG-C8-4-ABP, dG-C8-PhIP and dG-N2-B[a]DPE were spiked into 10 µg CT DNA digested directly while those of O6-Me-dG and O6-POB-dG were added to 5 µg CT DNA digest after adenosine deaminase and SPE clean-up. The levels of IS spiking are reported in Table S1. Each calibration point was assayed in triplicate. The molar ratios of the unlabeled adduct to its corresponding labeled adduct were: 0, 0.1, 0.5, 2.5, 5 (dG-C8-4-ABP); 0, 0.67, 3.33, 6.67, 33.3 (dG-C8-PhIP); 0, 0.04, 0.2, 1, 2 (dG-N2-B[a]PDE); 0, 0.05, 0.25, 0.5, 2, (O6-Me-dG) and 0, 0.1, 1, 2, 4 (O6-POB-dG). Data were fitted to a straight line (area of response of unlabeled/labeled adduct standard versus the amount of unlabeled/labeled adduct standard) using ordinary least-squares with equal weighting.
Method Validation
The accuracy of the method was validated with carcinogen-treated CT DNA containing known amounts of dG-C8-4-ABP, dG-N2-B[a]PDE, and dG-C8-PhIP. These DNA samples were provided by Dr. Frederick A. Beland from the National Center for Toxicology Research, US FDA.24,40,45 Kidney DNA of mice treated with AA-I (0.1 mg/kg) and previously assayed by UPLC-ESI-IT-MS3 was used to measure dA-AL-I.14,15 CT DNA alkylated with N-nitrosomethylurea and containing a known level of O6-Me-dG was provided by Dr. Lisa A. Peterson from University of Minnesota. DNA with a known level of O6-POB-dG was not available for assay. The within-day and between-day precision of the assay was further established by analyzing adduct levels in liver from the carcinogen-dosed rodent. Due to the limited quantity of DNA samples, fresh frozen or FFPE liver tissues of all animals from the same dosing experiment were homogenized and pooled. The methods of DNA retrieval, enzymatic digestion and UPLC-ESI-IT-MS3 described in the previous sections were employed. The % coefficient of variation (%CV) was used as the measure of precision.
RESULTS AND DISCUSSION
Method Validation
We adopted the UPLC-ESI-IT-MS3 method used previously for the measurement of aristolactam- DNA adducts to quantify dG-C8-4-ABP, dG-N2-B[a]PDE and dG-C8-PhIP, employing an online trap column to enrich analytes from the non-modified deoxynucleosides.14 However, O6-Me-dG and O6-POB-dG are hydrophilic and are not retained on the trap column. Moreover, a significant portion of dA was co-extracted with O6-Me-dG and O6-POB-dG by SPE, resulting in poor sensitivity for the ion trap MS. Thus, in order to remove the large excess of unmodified deoxynucleosides, particularly dA, adenosine deaminase was employed to convert dA into the more polar dI, which was efficiently removed by SPE. O6-Me-dG and O6-POB-dG were quantified by direct injection mode using the above UPLC-ESI-IT-MS3 method.
The robustness and reproducibility of the methods to measure the DNA adducts of these environmental and dietary carcinogens were validated with CT DNA modified in vitro with 4-ABP, PhIP and B[a]P,24,40,45 or methylated with N-nitrosomethylurea, and in vivo with mice treated with AA-I (0.1 mg/kg BW).14,15 Thereafter, the performance of the analytical method was evaluated with DNA retrieved from fresh frozen or FFPE liver tissues of mice treated with these carcinogens. All DNA adducts were measured in quadruplicate on three different days spanning a period of one week. Results of these studies are summarized in Table 1. The within-day and between-day precision values reported as %CV were within 19%. The accuracy values ranged between 0.82 and 1.36 of the targeted adduct values. The five calibration curves are shown in Figure S1. The linearity of the calibration curves are shown by the slope and the goodness-of-fit linear regression value (r2) were all higher than 0.998, and supported by residuals plots (data not shown). The calibration curves showed linearity over a 50-fold range of adduct levels.
Table 1.
Performance of Analytical Method to Measure DNA Adducts (per 107 nucleotides).
| Adduct | Accuracy (CT DNA modified with carcinogens) | In vivo (rodent tissue) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Target Value |
%CV |
Fresh Frozen Liver | %CV |
FFPE Liver | %CV |
||||||||||||
| Day 1 |
Day 2 |
Day 3 |
inter -day |
intra -day |
Day 1 |
Day 2 |
Day 3 |
inter -day |
intra -day |
Day 1 |
Day 2 |
Day 3 |
inter -day |
intra -day |
|||
| dG-N2- B[a]PDE |
Mean | 16 | 17 | 18 | 17 | 12 | 13 | 12 | 12 | 10 | 10 | ||||||
| SD | 1.0 | 1.5 | 1.2 | 1.4 | 0.81 | 1.3 | 1.6 | 0.48 | 1.1 | ||||||||
| %CV | 5.8 | 8.4 | 7.1 | 7.3 | 7.2 | 12 | 6.5 | 11 | 10 | 9.9 | 14 | 4.7 | 11 | 11 | 13 | ||
|
O6-Me- dG |
Mean | 10 | 14 | 13 | 14 | 152 | 147 | 150 | 222 | 199 | 180 | ||||||
| SD | 1.4 | 1.8 | 2.2 | 24 | 9.8 | 2.3 | 4.0 | 26 | 26 | ||||||||
| %CV | 10 | 14 | 16 | 14 | 13 | 16 | 6.6 | 2.0 | 10 | 9.5 | 1.8 | 13 | 14 | 10 | 13 | ||
|
O6-POB- dG |
Mean | 8.2 | 8.5 | 8.3 | 8.2 | 8.5 | 8.3 | 7.1 | 6.0 | 6.2 | |||||||
| SD | 1.2 | 0.77 | 0.66 | 1.2 | 0.77 | 0.66 | 0.47 | 0.39 | 0.82 | ||||||||
| %CV | 15 | 9.0 | 8.0 | 11 | 10 | 15 | 9.0 | 8.0 | 11 | 10 | 6.6 | 6.6 | 13 | 9.1 | 12 | ||
| dG-C8-4- ABP |
Mean | 1.9 | 1.6 | 1.6 | 1.5 | 15 | 15 | 15 | 22 | 22 | 22 | ||||||
| SD | 0.06 | 0.12 | 0.18 | 1.6 | 1.3 | 0.69 | 1.9 | 3.7 | 2.4 | ||||||||
| %CV | 3.7 | 7.2 | 12 | 7.9 | 8.1 | 10 | 8.6 | 4.7 | 8.3 | 7.7 | 8.4 | 17 | 11 | 12 | 11 | ||
| dG-C8- PhIP |
Mean | 1.0 | 1.1 | 1.2 | 1.1 | 0.42 | 0.41 | 0.41 | 0.25 | 0.30 | 0.31 | ||||||
| SD | 0.10 | 0.06 | 0.14 | 0.015 | 0.013 | 0.013 | 0.040 | 0.028 | 0.011 | ||||||||
| %CV | 8.7 | 5.3 | 13 | 9.4 | 9.0 | 3.4 | 3.1 | 3.1 | 3.2 | 3.6 | 16 | 9.3 | 3.6 | 10 | 14 | ||
| dA-AL-Ia | Mean | 5.0 | 6.9 | 5.2 | 5.0 | 1.8 | 1.5 | 1.7 | 1.5 | 1.5 | 1.4 | ||||||
| SD | 0.19 | 0.51 | 0.28 | 0.15 | 0.18 | 0.020 | 0.057 | 0.20 | 0.038 | ||||||||
| %CV | 2.8 | 9.7 | 5.6 | 6.3 | 19 | 8.0 | 12 | 1.1 | 8.0 | 12 | 3.9 | 13 | 2.8 | 8.2 | 9.7 | ||
Kidney DNA of mice treated with AA-I (0.1 mg/kg) was used to perform the accuracy measurement of dA-AL-I.
DNA Recovery from FFPE Tissues and the Stability of DNA Adducts
The aristolactam (AL-I)-DNA adduct dA-AL-I was successfully retrieved in high yield from FFPE tissues of rodents and from human tissues stored for up to nine years prior to analysis.15,46 Data from our rodent studies showed that even though the yield of DNA isolated from FFPE tissue decreased over time, following formalin fixation, the level of dA-AL-I remained constant.46 AA is one of the many diet-related and environmental-associated carcinogens that form DNA adducts in the genome. We sought to determine whether our DNA retrieval method could be used to recover, quantitatively, DNA adducts of other carcinogens from FFPE tissues. The major differences between the DNA retrieval methods for fresh frozen and FFPE tissues, besides the proprietary ingredients in the digestion and lysis buffers of the ZR FFPE DNA MiniPrep™ Kit, are the proteinase K digestion conditions: 37 °C for 2 h for fresh frozen tissue versus 50 °C overnight for FFPE tissue, and the DNA isolation methods, employing either phenol/chloroform extraction for fresh-frozen tissue or a silica-based extraction for FFPE tissue. The complete reversal of DNA cross-links required overnight (18 hr) proteolytic digestion of FFPE samples.
Key questions to address in the retrieval of DNA from FFPE tissue are: 1) Are other carcinogen DNA adducts stable towards formalin fixation and DNA extraction-digestion processes and can these adducts be quantified? 2) Does the formalin fixation process or the retrieval method introduce artifacts, such as aerial oxidation and ring-opening of the dG-C8-arylamine and HAA adducts?42,47
We conducted dosing experiments similar to those reported in the literature with 4-ABP, NNK, B[a]P, and AA-I in mouse models,15,25,35,48 and PhIP in rats.49 The major adducts formed in tissues of rodent tissues, prepared by either snap-freezing or formalin fixation were measured by UPLC-ESI-IT-MS3. The stabilities and levels of dG-C8-4-ABP, dG-N2-B[a]PDE and dG-C8-PhIP adducts recovered following digestion of DNA isolated by phenol/chloroform method with proteinase K at 37 °C for 2 h versus 50 °C overnight were determined. No major differences of the adduct level were observed between the two incubation conditions (Figure S2). Our previous data showed that dA-AL-I was also stable to overnight digestion at 50 °C.15 Some arylamine adducts formed at the C8 position of dG are prone to oxidation and undergo ring-opening to form spirohydantoin and guanidine adducts.42,47 The permeation of formalin through cell membranes causes oxidation of DNA and strand cleavage.46,50,51 Thus, we monitored formation of ring-opened adducts for dG-C8-4-ABP and dG-C8-PhIP from fresh frozen and FFPE tissues. Figure S3a and S3b show the EIC and MS3 scan stage product ion spectra of guanidine-, spirohydantoin-, dG-C8-4-ABP and dG-C8-PhIP adducts from the positive control DNA and from DNA extracted from carcinogen-treated rodent tissues, fresh-frozen and FFPE. Our findings demonstrate that dG-C8 adducts of 4-ABP and PhIP are stable towards the FFPE processing and DNA retrieval conditions.
Quantification of DNA Adducts in Rodent Tissues
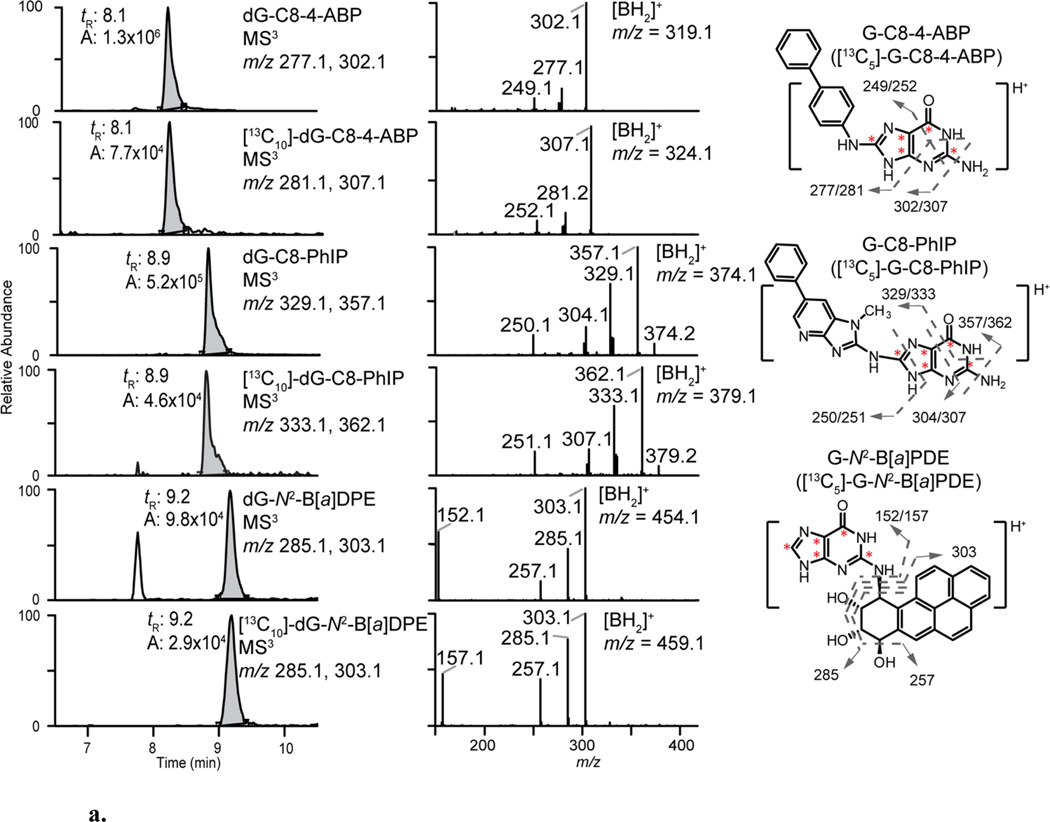
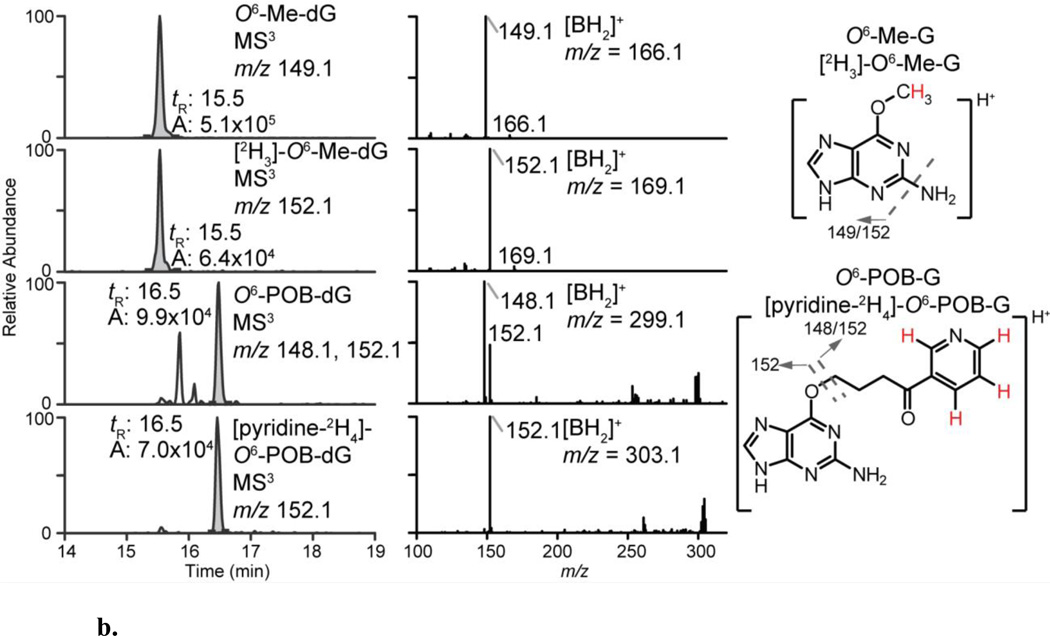
The EIC and product ion spectra of dG-C8-4-ABP, dG-C8-PhIP, dG-N2-B[a]PDE, O6-Me-dG, O6-POB-dG and their internal standards at the MS3 scan stage are depicted in Figure 2, while those data previously reported for dA-AL-I are shown in Figure S4. The levels of adducts from each animal group are shown as scatter plots in Figure 3. The %CV of adduct level measurement in each animal tissues, fresh-frozen or FFPE, were within 20% of the mean level (Table S3). The data on targeted adduct formation of B[a]P, 4-ABP, PhIP, NNK and AA-I previously performed by MS in other laboratories are summarized together with our new data in Table S2.
Figure 2.
a. EIC and product ion spectra of the adducts at the MS3 scan stage from DNA digest of carcinogen-treated rodent tissues: bladder for 4-ABP adducts; pancreas for PhIP adducts and lung, B[a]P. The m/z of aglycones ([BH2]+ = [M+H-116]+) are: G-C8-4-ABP (m/z 319.1), G-C8-PhIP (m/z 374.1), G-N2-B[a]PDE (m/z 454.1) and their internal standards (Δm/z = 5). The signature MS3 fragment ions of each adduct are shown on the right. The positions of 13C are indicated by asterisk symbols in red.
b. EIC and product ion spectra of the adducts at the MS3 scan stage from DNA digest of NNK-treated mouse liver. The m/z of aglycones ([BH2]+ = [M+H-116]+) are: O6-Me-G (m/z 166.1), O6-POB-G (m/z 299.1) and their internal standards (m/z 169.1 and 303.1, respectively). The signature MS3 fragment ions of each adduct are shown on the right. The positions of deuterium are highlighted in red.
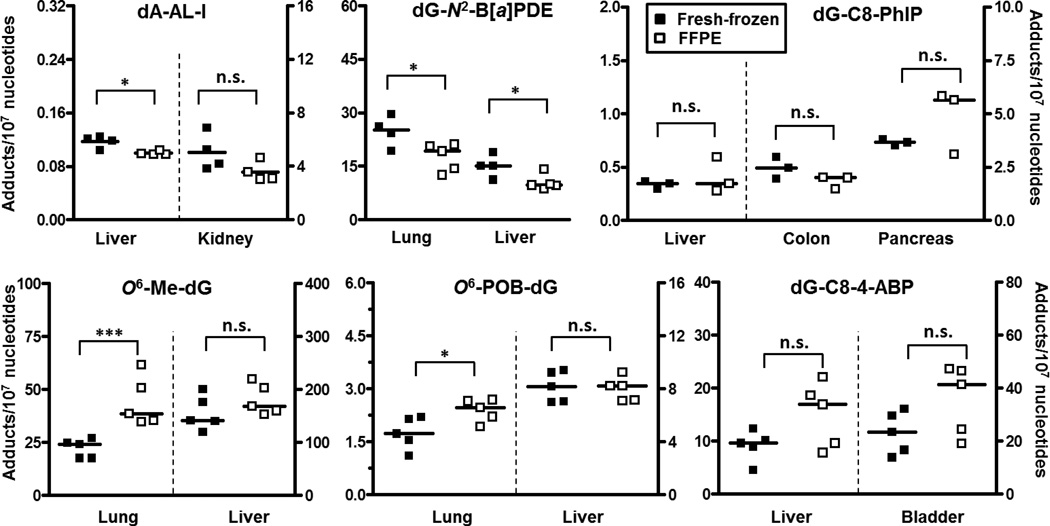
Figure 3.
A scatter plot with the median of DNA adduct levels measured from the carcinogen treated rodent tissues prepared in both fresh-frozen and FFPE method. Unpaired t-test (Prism 6, San Diego, CA): n.s., not significant; *p < 0.05; **p < 0.01; ***p < 0.005.
The mean levels of dG-C8-PhIP, dG-C8-4-ABP, hepatic O6-Me-dG and O6-POB-dG in fresh-frozen and FFPE tissue groups are not significantly different. However, the mean levels of dA-AL-I, dG-N2-B[a]PDE, pulmonary O6-Me-dG and O6-POB-dG in the fresh frozen and FFPE preserved tissues differed by up to 1.5-fold, and the tendency was for higher adduct levels in FFPE tissues. We attribute these differences in adduct levels to differences in the bioavailability of the test compound or in carcinogen metabolism among animals, We attribute these differences in adduct levels to differences in the bioavailability of the test compound or in carcinogen metabolism among animals, resulting in adduct levels that can vary by several fold,25,35,49 and not to methodological procedures, as our between-day %CV in DNA adduct measurements were on average <12%. The DNA digests were also assayed by HPLC-UV to confirm the purity and completeness of the digestion of DNA.15
Considering the large differences in assay conditions among laboratories, including the employment of different internal standards, enzymes and DNA digestion conditions, and MS instrumentation, the levels of hepatic dG-N2-B[a]PDE, dG-C8-4-ABP, pulmonary O6-Me-dG, O6-POB-dG adduct levels measured in our study are quite comparable to previously reported values by other laboratories employing the same dosages of carcinogens (Table S2).25,35,48,52 For pulmonary dG-N2-B[a]PDE, bladder dG-C8-4-ABP, hepatic O6-Me-dG, O6-POB-dG, no direct comparison can be drawn as adduct levels are not reported in the literature. We observed similar carcinogenic DNA adduct distribution among different organs within the same animals as previously reported. For example, NNK formed the dG adducts O6-Me-dG and O6-POB-dG at approximately 4-fold greater levels in liver than lung.36,53 The amount of the dG-C8-4-ABP was two-fold greater in bladder than liver of mice and consistent with etiologic role of 4-ABP in bladder cancer.54 In PhIP-treated rats, we observed the same trend in the level of dG-C8-PhIP adduct formation as previously reported, with highest levels of adducts formed in the pancreas followed by colon, and then liver.49,55
CONCLUSIONS
The findings reported here demonstrate that the major DNA adducts formed from B[a]P, 4-ABP, PhIP, NNK and AA-I, representing five important classes of environmental and dietary carcinogens, can be retrieved from FFPE tissues of rodents in high yield and quantified by UPLC-ESI-IT-MS3. The levels of DNA adducts measured in fresh-frozen and FFPE tissues of liver, lung, bladder, pancreas, and colon of rodents were comparable. Adducts targeted in this study cover both hydrophobic adducts, such as dG-C8-4-ABP and dG-N2-B[a]PDE, and hydrophilic adducts, such as O6-Me-dG. The use of adenosine deaminase and offline DNA adduct enrichment by SPE to eliminate dA was necessary for the measurement of O6-Me-dG. However, ~20% of O6-Me-dG in the DNA digest was demethylated by adenosine deaminase (unpublished studies, J. Guo). Adenine deaminase possesses O6-demethylase activity,44 and the hydrolysis of various groups at the 6-position of purine ribonucleosides, including 6-methoxyguanosine, by adenosine deaminase, has been reported by Baer and coworkers.44 However, adenosine deaminase treatment had no effect on the level of O6-POB-dG (unpublished studies, J. Guo). Thus, careful optimization of post-DNA digestion with adenine deaminase is required for the measurements of other O6 alkylated dG adducts. All adducts analyzed in this study were found to be stable during FFPE processing and DNA retrieval.The analytical data demonstrate that the methodology originally established for biomonitoring adducts of AA-I in FFPE tissues15,46 can be used to screen for other classes of environmental and dietary genotoxicants, in molecular epidemiology studies designed to assess the causal role of exposures to hazardous chemicals in cancer risk.
Supplementary Material
Acknowledgments
We thank Dr. Frederick A. Beland from the National Center for Toxicology Research /US FDA for generously providing us the B[a]P, 4-ABP and PhIP-treated CT DNA, and Dr. Lisa Peterson from University of Minnesota for providing CT DNA samples containing O6-Me-dG.
Funding: Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number R33CA186795 (R.J.T.) and in part by and Cancer Center Support grant no. CA077598 (to R.J.T.)
REFERENCES
- 1.Sugimura T. Toxicology. 2002;181–182:17–21. doi: 10.1016/s0300-483x(02)00250-0. [DOI] [PubMed] [Google Scholar]
- 2.Dedon PC, Tannenbaum SR. Arch. Biochem. Biophys. 2004;423:12–22. doi: 10.1016/j.abb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 3.Miller EC. Cancer Res. 1978;38:1479–1496. [PubMed] [Google Scholar]
- 4.Himmelstein MW, Boogaard PJ, Cadet J, Farmer PB, Kim JH, Martin EA, Persaud R, Shuker DE. Crit. Rev.Toxicol. 2009;39:679–694. doi: 10.1080/10408440903164163. [DOI] [PubMed] [Google Scholar]
- 5.Jarabek AM, Pottenger LH, Andrews LS, Casciano D, Embry MR, Kim JH, Preston RJ, Reddy MV, Schoeny R, Shuker D, Skare J, Swenberg J, Williams GM, Zeiger E. Crit.Rev.Toxicol. 2009;39:659–678. doi: 10.1080/10408440903164155. [DOI] [PubMed] [Google Scholar]
- 6.Gavina JMA, Yao CH, Feng YL. Talanta. 2014;130:475–494. doi: 10.1016/j.talanta.2014.06.050. [DOI] [PubMed] [Google Scholar]
- 7.Poirier MC, Santella RM, Weston A. Carcinogenesis. 2000;21:353–359. doi: 10.1093/carcin/21.3.353. [DOI] [PubMed] [Google Scholar]
- 8.Phillips DH. Cancer Lett. 2012;334:5–9. doi: 10.1016/j.canlet.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 9.Fox CH, Johnson FB, Whiting J, Roller PP. J. Histochem. Cytochem. 1985;33:845–853. doi: 10.1177/33.8.3894502. [DOI] [PubMed] [Google Scholar]
- 10.Nirmalan NJ, Harnden P, Selby PJ, Banks RE. Mol.Biosyst. 2008;4:712–720. doi: 10.1039/b800098k. [DOI] [PubMed] [Google Scholar]
- 11.Lewis F, Maughan NJ, Smith V, Hillan K, Quirke P. J. Pathol. 2001;195:66–71. doi: 10.1002/1096-9896(200109)195:1<66::AID-PATH921>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 12.Klopfleisch R, Weiss AT, Gruber AD. Histol Histopathol. 2011;26:797–810. doi: 10.14670/HH-26.797. [DOI] [PubMed] [Google Scholar]
- 13.Grollman AP. Environ. Mol. Mutagen. 2013;54:1–7. doi: 10.1002/em.21756. [DOI] [PubMed] [Google Scholar]
- 14.Yun BH, Rosenquist TA, Sidorenko V, Iden CR, Chen CH, Pu YS, Bonala R, Johnson F, Dickman KG, Grollman AP, Turesky RJ. Chem. Res. Toxicol. 2012;25:1119–1131. doi: 10.1021/tx3000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yun BH, Rosenquist TA, Nikolic J, Dragicevic D, Tomic K, Jelakovic B, Dickman KG, Grollman AP, Turesky RJ. Anal. Chem. 2013;85:4251–4258. doi: 10.1021/ac400612x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jelakovic B, Karanovic S, Vukovic-Lela I, Miller F, Edwards KL, Nikolic J, Tomic K, Slade N, Brdar B, Turesky RJ, Stipancic Z, Dittrich D, Grollman AP, Dickman KG. Kidney Int. 2012;81:559–567. doi: 10.1038/ki.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanherweghem JL, Debelle FD, Muniz Martinez MC, Nortier JL. In: Clinical Nephrotoxins. 2nd. De Broe ME, Porter GA, Bennett WM, Verpooten GA, editors. Dordrecht: Kluwer; 2003. pp. 579–603. [Google Scholar]
- 18.De Broe ME. Kidney Int. 2012;81:513–515. doi: 10.1038/ki.2011.428. [DOI] [PubMed] [Google Scholar]
- 19.Hoang ML, Chen CH, Sidorenko VS, He J, Dickman KG, Yun BH, Moriya M, Niknafs N, Douville C, Karchin R, Turesky RJ, Pu YS, Vogelstein B, Papadopoulos N, Grollman AP, Kinzler KW, Rosenquist TA. Sci. Transl. Med. 2013;5:197ra102. doi: 10.1126/scitranslmed.3006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phillips DH. Mutat. Res. 1999;443:139–147. doi: 10.1016/s1383-5742(99)00016-2. [DOI] [PubMed] [Google Scholar]
- 21.IInternational Agency for Research on Cancer. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans: Tobacco smoking. Vol. 38. France: Lyon; 1986. [PMC free article] [PubMed] [Google Scholar]
- 22.Felton JS, Jagerstad M, Knize MG, Skog K, Wakabayashi K. In: Food Borne Carcinogens Heterocyclic Amines. Nagao M, Sugimura T, editors. Chichester, England: John Wiley & Sons Ltd; 2000. pp. 31–71. [Google Scholar]
- 23.International Agency for Research on Cancer. Some aromatic amines, organic dyes, and related exposures. Vol. 99. France: Lyon; 2010. IARC monographs on the evaluation of carcinogenic risks to humans. [PMC free article] [PubMed] [Google Scholar]
- 24.Beland FA, Churchwell MI, Von Tungeln LS, Chen S, Fu PP, Culp SJ, Schoket B, Gyorffy E, Minarovits J, Poirier MC, Bowman ED, Weston A, Doerge DR. Chem. Res. Toxicol. 2005;18:1306–1315. doi: 10.1021/tx050068y. [DOI] [PubMed] [Google Scholar]
- 25.Singh R, Gaskell M, Le Pla RC, Kaur B, Azim-Araghi A, Roach J, Koukouves G, Souliotis VL, Kyrtopoulos SA, Farmer PB. Chem. Res. Toxicol. 2006;19:868–878. doi: 10.1021/tx060011r. [DOI] [PubMed] [Google Scholar]
- 26.Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. Oncogene. 2002;21:7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 27.Turesky RJ, Freeman JP, Holland RD, Nestorick DM, Miller DW, Ratnasinghe DL, Kadlubar FF. Chem. Res. Toxicol. 2003;16:1162–1173. doi: 10.1021/tx030029r. [DOI] [PubMed] [Google Scholar]
- 28.Neumann HG. Chemosphere. 2001;42:473–479. doi: 10.1016/s0045-6535(00)00219-8. [DOI] [PubMed] [Google Scholar]
- 29.Ni W, McNaughton L, LeMaster DM, Sinha R, Turesky RJ. J. Agric. Food Chem. 2008;56:68–78. doi: 10.1021/jf072461a. [DOI] [PubMed] [Google Scholar]
- 30.Zheng W, Lee SA. Nutr. Cancer. 2009;61:437–446. doi: 10.1080/01635580802710741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beland FA, Beranek DT, Dooley KL, Heflich RH, Kadlubar FF. Environ. Health Perspect. 1983;49:125–134. doi: 10.1289/ehp.8349125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bessette EE, Goodenough AK, Langouet S, Yasa I, Kozekov ID, Spivack SD, Turesky RJ. Anal. Chem. 2009;81:809–819. doi: 10.1021/ac802096p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.International Agency for Research on Cancer. IARC Monograph on the Smokeless Tobacco and Tobacco-Specific Nitrosamines. Vol. 89. France: Lyon; 2007. [Google Scholar]
- 34.Hecht SS. Chem. Res. Toxicol. 2008;21:160–171. doi: 10.1021/tx7002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson LA, Hecht SS. Cancer Res. 1991;51:5557–5564. [PubMed] [Google Scholar]
- 36.Upadhyaya P, Lindgren BR, Hecht SS. Drug Metab. Dispos. 2009;37:1147–1151. doi: 10.1124/dmd.109.027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hecht SS. Chem. Res. Toxicol. 1998;11:559–603. doi: 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- 38.Hecht SS, Lin D, Castonguay A. Carcinogenesis. 1983;4:305–310. doi: 10.1093/carcin/4.3.305. [DOI] [PubMed] [Google Scholar]
- 39.Lao Y, Villalta PW, Sturla SJ, Wang M, Hecht SS. Chem. Res. Toxicol. 2006;19:674–682. doi: 10.1021/tx050351x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin D, Kaderlik KR, Turesky RJ, Miller DW, Lay JO, Jr, Kadlubar FF. Chem. Res. Toxicol. 1992;5:691–697. doi: 10.1021/tx00029a016. [DOI] [PubMed] [Google Scholar]
- 41.Weng Y, Fang C, Turesky RJ, Behr M, Kaminsky LS, Ding X. Cancer Res. 2007;67:7825–7832. doi: 10.1158/0008-5472.CAN-07-1006. [DOI] [PubMed] [Google Scholar]
- 42.Tang Y, Kassie F, Qian X, Ansha B, Turesky RJ. Toxicol. Sci. 2013;133:248–258. doi: 10.1093/toxsci/kft077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nauwelaers G, Bessette EE, Gu D, Tang Y, Rageul J, Fessard V, Yuan JM, Yu MC, Langouët S, Turesky RJ. Chem. Res. Toxicol. 2011;24:913–925. doi: 10.1021/tx200091y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baer HP, Drummond GI, Gillis J. Arch. Biochem. Biophys. 1968;123:172–178. doi: 10.1016/0003-9861(68)90116-1. [DOI] [PubMed] [Google Scholar]
- 45.Beland FA, Doerge DR, Churchwell MI, Poirier MC, Schoket B, Marques MM. Chem. Res. Toxicol. 1999;12:68–77. doi: 10.1021/tx980172y. [DOI] [PubMed] [Google Scholar]
- 46.Yun BH, Yao L, Jelakovic B, Nikolic J, Dickman KG, Grollman AP, Rosenquist TA, Turesky RJ. Carcinogenesis. 2014;35:2055–2061. doi: 10.1093/carcin/bgu101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shibutani S, Gentles RG, Iden CR, Johnson F. J. Am. Chem. Soc. 1990;112:5667–5668. [Google Scholar]
- 48.Doerge DR, Churchwell MI, Marques MM, Beland FA. Carcinogenesis. 1999;20:1055–1061. doi: 10.1093/carcin/20.6.1055. [DOI] [PubMed] [Google Scholar]
- 49.Friesen MD, Kaderlik K, Lin D, Garren L, Bartsch H, Lang NP, Kadlubar FF. Chem. Res. Toxicol. 1994;7:733–739. doi: 10.1021/tx00042a004. [DOI] [PubMed] [Google Scholar]
- 50.Gilbert MT, Haselkorn T, Bunce M, Sanchez JJ, Lucas SB, Jewell LD, Van ME, Worobey M. PLoS One. 2007;2:e537. doi: 10.1371/journal.pone.0000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peluso EM, Munnia A, Tarocchi M, Giese RW, Annaratone L, Bussolati G, Bono R. Toxicol. Res. 2014;3:341–349. [Google Scholar]
- 52.Urban AM, Upadhyaya P, Cao Q, Peterson LA. Chem. Res. Toxicol. 2012;25:2167–2178. doi: 10.1021/tx300245w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sandercock LE, Hahn JN, Li L, Luchman HA, Giesbrecht JL, Peterson LA, Jirik FR. Carcinogenesis. 2008;29:866–874. doi: 10.1093/carcin/bgn030. [DOI] [PubMed] [Google Scholar]
- 54.Poirier MC, Beland FA. Chem. Res. Toxicol. 1992;5:749–755. doi: 10.1021/tx00030a003. [DOI] [PubMed] [Google Scholar]
- 55.Goodenough AK, Schut HA, Turesky RJ. Chem. Res. Toxicol. 2007;20:263–276. doi: 10.1021/tx0601713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.