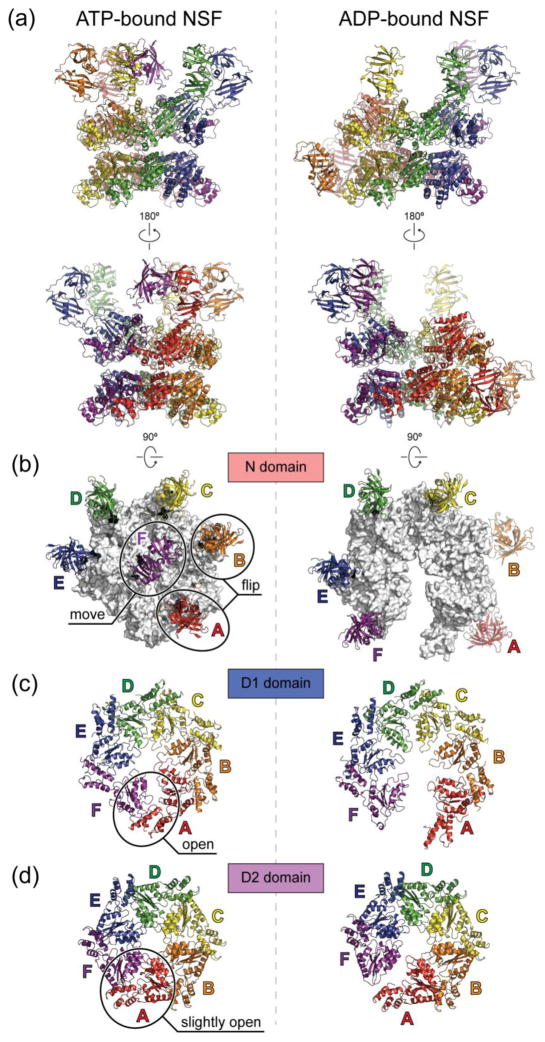
Fig. 4. Cryo-EM structures of ATP- and ADP-bound full-length NSF.
The cryo-EM structures are derived from PDB entries 3j94 and 3j95 with the N domains docked into the density of unsharpened maps of ATP- and ADP-bound NSF respectively as rigid bodies, using the crystal structure of the N domain (PDB accession code: 1qcs). (a), Side views. The six chains are colored red, orange, yellow, green, blue, and purple counter-clockwise as viewed from the top. The red chain is always the one with the lowest α2 helix in the D1 domain. (b), Top views of the N domains. The D1 rings are colored white. (c), Top views of the D1 domains. (d), Top views of the D2 domains. Conformational changes upon ATP hydrolysis are highlighted in the ATP-bound structures on the left in panels (b–d). All structures are shown in scale.