Figure 1.
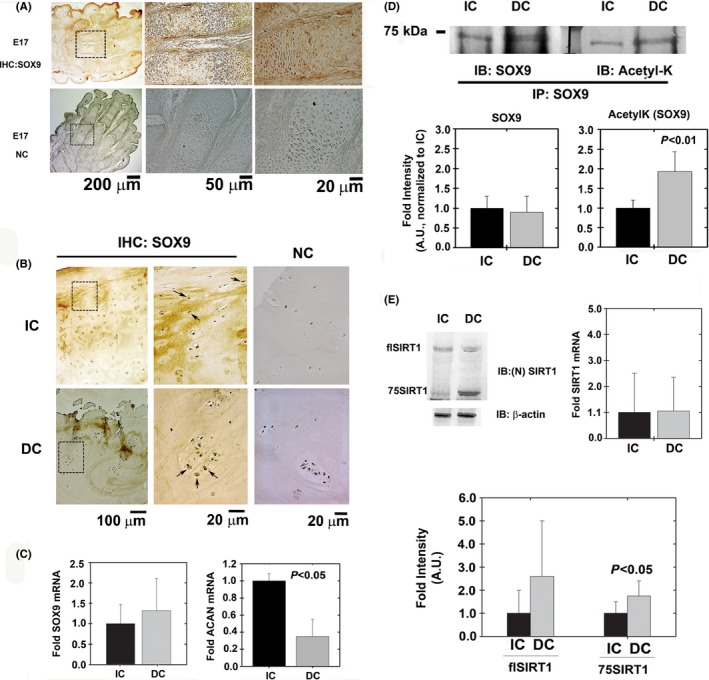
SOX9 levels in healthy and diseased articular cartilage. (A) Sagittal sections of E17 hind paws served as positive and negative controls (NC) for immunohistochemistry (IHC) protocols to detect SOX9 protein levels (n = 5).(B) OA‐derived intact cartilage (IC) and degenerated cartilage (DC) were analyzed for SOX9 levels using immunohistochemistry (n = 10). (C) SOX9 and ACAN mRNA levels of freshly isolated chondrocytes from IC and DC samples (n = 8). As indicated in the Materials and methods section, GAPDH was used as housekeeping gene. (D) SOX9 from IC and DC samples was immunoprecipitated and blotted for SOX9 and acetyl‐lysine (AcetylK; n = 5). (E) Freshly isolated chondrocytes from IC and DC cartilage were analyzed with an N‐terminally reactive antibody for SIRT1 to identify its cleavage (n = 9). Full‐length SIRT1 is denoted as flSIRT1, while 75SIRT1 is the 75 kDa cleaved variant generated by cathepsin B‐mediated cleavage of SIRT1 on its C‐terminal domain. Right graphs shows mRNA expression for SIRT1 in freshly isolated chondrocytes from IC and DC samples (n = 8). Image J was employed to quantify band intensity of all immunoblots. SOX9 acetylation was calculated based on the intensity of acetyl‐lysine (AcetylK) band divided by the band intensity of normalized SOX9, assuming the immunoprecipitate consists mainly of SOX9. Statistical significance was determined based on Mann–Whitney U‐test, assuming a P < 0.05 to be statistically significant.
