Figure 4.
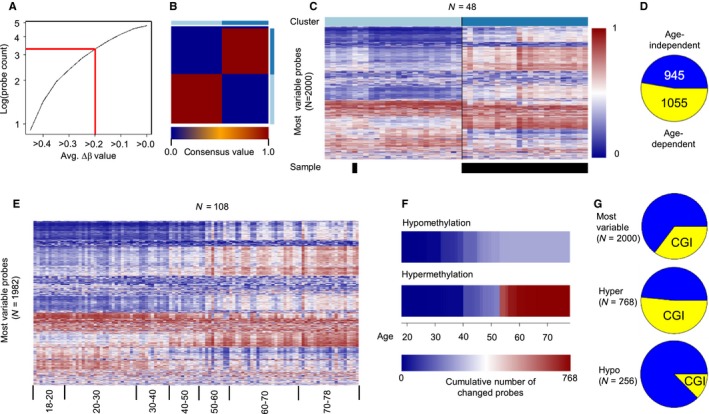
Discontinuous methylation changes during aging. (A) Number of differentially methylated probes in relation to their age‐related (young vs. old) average methylation difference. Approximately 2000 probes showed an average age‐related methylation difference of greater than or equal to 0.2. (B) Consensus Matrix for two cluster centers after consensus clustering of the 2000 most variable probes (young vs. old). Only consensus values of 0 (two samples never cluster together) or 1 (two samples always cluster together) were observed, indicating optimum clustering into two groups. (C) β value heatmap of the 2000 most variable probes. β values were colored from blue (β = 0) to red (β = 1). Colors in the bar above the matrix indicate cluster assignment. Epidermis methylomes from the old sample group are indicated by black boxes below the heatmap. Only one sample appeared in the wrong cluster. (D) Distribution of age‐dependent and age‐independent β values within the 2000 most variable probes. Yellow: age‐dependent probes; Blue: age‐independent probes. (E) β value heatmap of the most variable probes within the complete dataset after sorting by age. Discontinuous methylation changes occur for a subset of probes between the ages of 40 and 60. (F) Identification of probes showing discontinuous methylation changes by recursive partitioning. The heatmaps represent the cumulative number of probes showing a discontinuous β value change at the specified age. Hypomethylation occurred in less probes than hypermethylation and at different ages. (G) Fraction of CpG island‐associated probes (yellow) among the most variable (top), hypermethylated (middle) and hypomethylated (bottom) probes.
