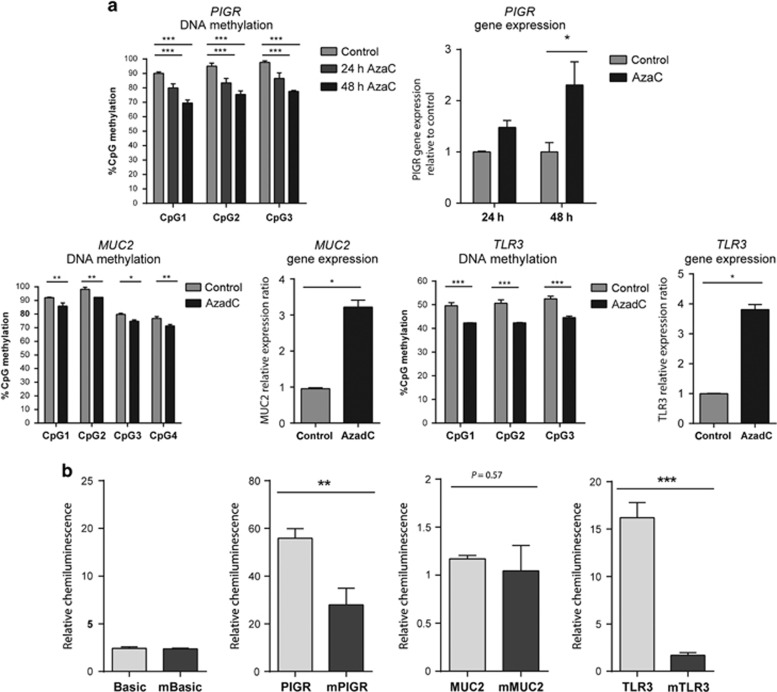
Figure 6.
In-vitro assays demonstrate the direct impact of DNA methylation on transcription of regulatory differentially methylated region (rDMR)-containing genes. (a) Caco-2 cells were treated with DNA methyltransferase (DNMT) inhibitor (10 μM 5-Azacytidine (AzaC) for 24 h and for 48 h or 5 mM 5-Azadeoxycytidine (AzadC) for 24 h. DNA methylation of the polymeric immunoglobulin receptor (PIGR), MUC2, and TLR3 promoter was assessed by pyrosequencing. Reduced levels of CpG methylation with a corresponding increase in mRNA expression was observed. Data are displayed as mean+s.d. of three independent experiments performed in duplicates. Two-way analysis of variance, post test; Sidak, *P<0.05, **P<0.005, ***P<0.001 vs. dimethylsulfoxide (DMSO) vehicle control. (b) Promoter regions of MUC2, TLR3, and PIGR were cloned into a CpG-free luciferase reporter plasmid. In-vitro methylation of TLR3 and PIGR promoter plasmids led to a significant reduction in luciferase signal compared with mock-treated unmethylated plasmids. Basic luciferase activity of MUC2 promoter containing plasmid was found to be low in Caco-2 cells and no further reduction was observed following in-vitro methylation. Basic, CpG-free plasmid with no promoter. Renilla-luciferase served as control to correct for transfection efficiency. Mean+s.d. of three independent experiments performed in duplicates; Student's unpaired t-test, two tailed, n=3 per condition. ***P<0.0001, **P<0.005, P<0.57 (NS, not significant).