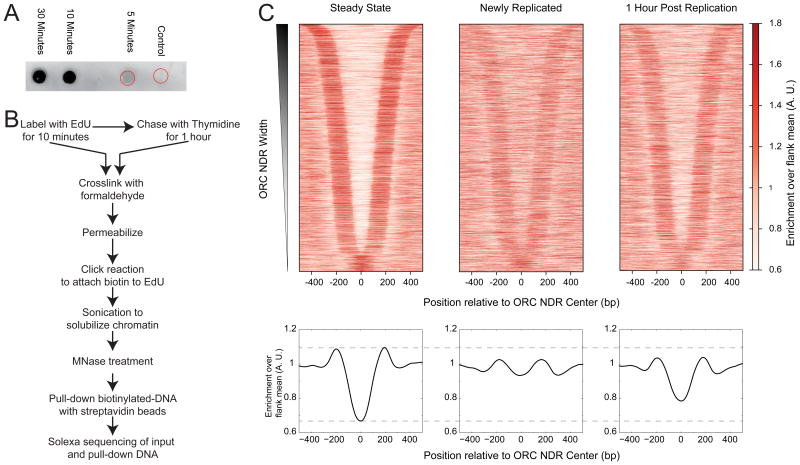
Figure 1. EdU labeling of newly replicated chromatin.
A) Dot blot of DNA purified from chromatin preparations of cells treated with EdU for indicated times. Control cells were not treated with EdU, but were processed the same way as other samples. Red circles indicate the location of sample application for control and 5 minute samples. The DNA applied to a nitrocellulose membrane was probed with IRDye 800CW streptavidin and detected using ODYSSEY infrared scanner. B) Schematic of the MINCE-seq protocol C) Nucleosome profiles of steady state, newly replicated and 1 hour post replication plotted as heatmaps (top) and average plots (bottom) around ORC nucleosome-depleted regions (NDRs; n=4230). NDRs were computationally identified around published ORC sites, and the center of the NDR was used as the reference point for plotting nucleosome profiles. All profiles are averages over a 50-bp sliding window. See also Figure S1 and Tables S1.