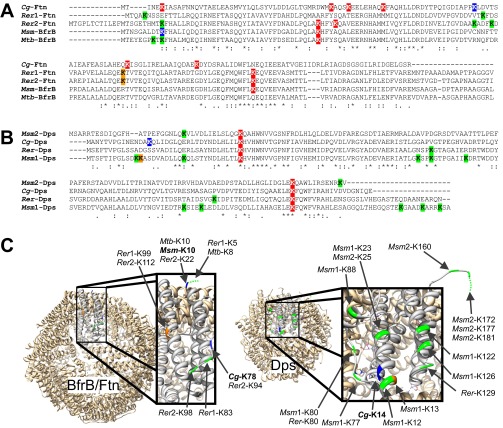
Fig. S8.
Pupylation sites in the iron storage proteins ferritin and Dps. To determine the spatial positions of identified and putative pupylation sites in ferritin and Dps, amino acid sequence alignments were made to homologs of known structure, BfrB from M. tuberculosis (Mtb-BfrB, Rv3841) and Dps from Mycobacterium smegmatis (Msm1-Dps, MSMEG_6467), using ClustalO (www.ebi.ac.uk/Tools/msa/clustalo/). (A) The ferritin sequences from C. glutamicum (Cg2782), Rhodococcus erythropolis (Rer1, RER_01810 und Rer2, RER_01740), and M. smegmatis BfrB (MSMEG_6422) were aligned to Mtb-BfrB. (B) The Dps sequences from M. smegmatis (Msm2, MSMEG_3242), C. glutamicum (Cg3327), and R. erythropolis (RER_12800) were aligned to Msm1-Dps. (C) Protein structures of the 24-mer BfrB from M. tuberculosis [Protein Data Bank (PDB)_ID code 3QD8] and the 12-mer Dps from M. smegmatis (PDB ID code 1VEQ) with one of the monomers enlarged. The 3D models were exported by using Chimera (55). Based on the alignments in A and B, the positions of all lysine residues were determined in the protein structures and are indicated in colors. Lysine residues confirmed as pupylation sites are colored blue, and lysine residues located on top of helices or in loop regions that could be easily accessible for pupylation are depicted in green. Lysine residues shown in orange are located at the flanks of helices, making accessibility less likely. Lysine residues not shown in C are likely facing the interior cave of the oligomers and therefore should be inaccessible for pupylation. Lysine residues colored red in A and B are confirmed (Cg-Ftn) or assumed (Cg-Dps) not to be pupylated in C. glutamicum or inaccessible for pupylation.