Significance
Penicillin-binding proteins (PBPs) are the targets of penicillin and related beta-lactams, one of our oldest and most effective classes of antibiotics. These enzymes assemble the essential bacterial cell wall. Their structure and biochemical activities have been well-characterized in vitro. However, surprisingly little is known about how PBP activity is controlled in cells. Here, we investigate the mechanism of PBP activation in Escherichia coli by a lipoprotein cofactor located in the outer membrane. Our results suggest that activation proceeds via a conformational change in the PBP induced by lipoprotein binding. This information, as well as the nature of the PBP variants isolated, provides new insight into the function of these important drug targets.
Keywords: peptidoglycan, cell wall, PBP, penicillin, murein
Abstract
To fortify their cytoplasmic membrane and protect it from osmotic rupture, most bacteria surround themselves with a peptidoglycan (PG) exoskeleton synthesized by the penicillin-binding proteins (PBPs). As their name implies, these proteins are the targets of penicillin and related antibiotics. We and others have shown that the PG synthases PBP1b and PBP1a of Escherichia coli require the outer membrane lipoproteins LpoA and LpoB, respectively, for their in vivo function. Although it has been demonstrated that LpoB activates the PG polymerization activity of PBP1b in vitro, the mechanism of activation and its physiological relevance have remained unclear. We therefore selected for variants of PBP1b (PBP1b*) that bypass the LpoB requirement for in vivo function, reasoning that they would shed light on LpoB function and its activation mechanism. Several of these PBP1b variants were isolated and displayed elevated polymerization activity in vitro, indicating that the activation of glycan polymer growth is indeed one of the relevant functions of LpoB in vivo. Moreover, the location of amino acid substitutions causing the bypass phenotype on the PBP1b structure support a model in which polymerization activation proceeds via the induction of a conformational change in PBP1b initiated by LpoB binding to its UB2H domain, followed by its transmission to the glycosyl transferase active site. Finally, phenotypic analysis of strains carrying a PBP1b* variant revealed that the PBP1b–LpoB complex is most likely not providing an important physical link between the inner and outer membranes at the division site, as has been previously proposed.
The peptidoglycan (PG) layer forms a protective shell that surrounds the cytoplasmic membrane of bacteria to prevent osmotic rupture and provide cells with their characteristic shape (1). This complex macromolecule is composed of glycan strands crosslinked to one another by attached peptide chains to form the exoskeletal matrix. Because of its essentiality and uniqueness to bacteria, the PG layer is an important therapeutic target. Many antibiotics in our current arsenal block PG assembly, with penicillin and related beta-lactam drugs being the most well-known and widely used. These molecules target major PG synthase enzymes called penicillin-binding proteins (PBPs) (1).
The PBPs function in the final stage of the three-part pathway for PG biogenesis (1). Precursor synthesis begins in the cytoplasm with the production of the activated sugar molecules uridine diphosphate (UDP)-N-acetylmuramic acid pentapeptide (UDP-MurNAc-pep5) and UDP-N-acetylglucosamine (UDP-GlcNAc). In the second, membrane-associated phase, UDP-MurNAc-pep5 is converted to the precursor lipid-I by MraY, which transfers phospho-MurNAc-pep to the lipid carrier undecaprenol-phosphate (Und-P). Lipid-II is formed by MurG via the addition of GlcNAc to lipid-I from UDP-GlcNAc. This final precursor contains the basic monomeric unit of PG, the disaccharide-pentapeptide. After its production, lipid-II must be flipped by MurJ (2, 3) to expose the sugar units to the periplasmic space, where it can then be polymerized and crosslinked into PG by the PBPs. The synthetic PBPs fall into two classes, A and B, on the basis of their enzymatic activities. Class A PBPs (aPBPs) are bifunctional enzymes that possess both PG glycosyltransferase (PGT) activity to polymerize glycan strands of PG from lipid-II and transpeptidase (TP) activity to generate the peptide crosslinks between adjacent glycans. The class B enzymes (bPBPs), in contrast, are only known to have TP activity (1).
To build the dynamic, cell-shaped PG matrix, the PBPs are thought to function in the context of multiprotein complexes that are organized by cytoskeletal elements (1). In rod-shaped bacteria, the Rod complex is organized by the actin-like protein MreB to promote the insertion of new PG material along the cell cylinder for cell elongation (1). After elongation, the tubulin-like FtsZ protein promotes the formation of the divisome (septal ring) machine to catalyze cell division and build the PG material that will ultimately fortify the new daughter cell poles (1). In Escherichia coli, the aPBPs, PBP1a, and PBP1b are thought to play important roles in both cell elongation and cell division. Localization and protein–protein interaction studies suggest that PBP1a primarily participates in cell elongation, whereas PBP1b mainly functions during cell division (4–6). However, neither aPBP is essential for growth, indicating they can likely substitute for one another and are not specific for a particular mode of growth.
Although PBP1a and PBP1b are individually dispensable, simultaneous inactivation of both factors results in rapid cell lysis and death (7, 8). We used this synthetic lethal phenotype to identify outer membrane lipoproteins that are required for the in vivo function of the aPBPs (9). LpoA is specifically required for PBP1a function, whereas LpoB is specifically required for PBP1b function (9, 10). Although they are unrelated, LpoA and LpoB both span the periplasm to interact directly with their cognate PBP (11–13), and when added to in vitro reactions, they modulate the PGT and TP activities of their target synthases via different mechanisms (9, 10, 14). LpoA activates the TP activity of PBP1a, which indirectly stimulates its PGT activity, indicating a coupling between the two active sites (10, 14). Similarly, LpoB stimulates the PGT activity of PBP1b, which in turn provides more material for crosslinking by the TP domain (9, 12). At present, the mechanism of PBP activation by the Lpo factors remains unclear. Also, because the activation has only been observed in vitro, its physiological relevance in vivo has yet to be established.
To investigate PBP activation further, we developed a genetic selection strategy to identify variants of PBP1b (referred to as PBP1b*) that bypass the requirement for LpoB. Several of these variants were purified and shown to have elevated PGT activity in vitro, indicating that PGT activation is indeed one of the relevant functions of LpoB in vivo. Moreover, the location of the amino acid substitutions in the PBP1b* bypass variants support a model in which PGT activation proceeds via the induction of a conformational change in PBP1b initiated by LpoB binding to the UB2H domain of the enzyme, followed by its transmission to the PGT active site. Finally, phenotypic analysis of strains carrying a PBP1b* variant revealed that the PBP1b–LpoB complex is most likely not providing an important physical link between the inner and outer membranes at the division site, as has been previously proposed (10).
Results
Identification of PBP1b* Variants That Bypass the LpoB Requirement for in Vivo Activity.
To identify variants of PBP1b that no longer require LpoB (designated PBP1b*), we used the cefsulodin-hypersensitivity phenotype of mutants inactivated for PBP1b (ΔponB) or LpoB (ΔlpoB) (9, 10). Cefsulodin is a β-lactam that preferentially targets PBP1a and PBP1b in E. coli (15). We reasoned that the cefsulodin hypersensitivity of ΔlpoB mutants likely results from a defect in PBP1b activity and that ponB* alleles encoding PBP1b* variants might be identified by a selection for increased cefsulodin resistance in a LpoB− strain background. Therefore, cells of MM33(attλTB309) [ΔlpoB ΔponA (Para::ponA)] were plated at 30 °C on M9-arabinose agar supplemented with 0.01 µg/mL cefsulodin for the isolation of spontaneous, drug-resistant mutants. Survivors arose at a frequency of ∼10−7. In addition to the ponB* alleles we sought, we anticipated a background of mutants encoding PBP1a variants with a reduced sensitivity to cefsulodin or mutants overproducing resistance pumps. To rapidly identify ponB* mutants among this expected background, we used the arabinose-dependent growth phenotype of MM33(attλTB309) [ΔlpoB ΔponA (Para::ponA)], which results from the simultaneous inactivation of LpoB and PBP1a (9, 10). Cefsulodin-resistant survivors were therefore screened for growth in the absence of PBP1a induction. Survivors producing cefsulodin-resistant PBP1a variants or overproducing efflux pumps are predicted to be arabinose-dependent for growth because PBP1a activity will remain essential in the absence of LpoB-stimulated PBP1b activity. Mutants producing PBP1b* variants, in contrast, should grow without arabinose because the PBP1b synthetic pathway will be functional without LpoB, thus suppressing the synthetic lethality of the PBP1a− LpoB− combination.
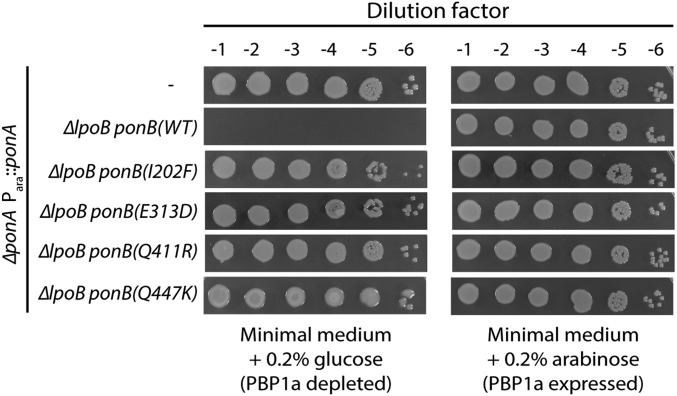
We isolated ∼600 mutants resistant to 0.01 µg/mL cefsulodin. Among these, 16 arabinose-independent mutants were identified, bringing the overall frequency of mutant isolation to ∼10−9, which reflects the requirement for specific missense mutations to allow LpoB bypass. This phenotype was ∼50% linked to a transposon insertion in yadC (yadC::Tn10) (16), located about 12 kb upstream of the ponB gene. We therefore sequenced the ponB gene from all the arabinose-independent isolates. In all cases, the reading frame encoded PBP1b variants with one of five amino acid substitutions (E313D, I202F, Q411R, Q411K, and Q447K) (Fig. 1 and SI Appendix, Table S1). Interestingly, the mutants producing PBP1b(I202F) and PBP1b(Q447K) displayed temperature-sensitive bypass phenotypes, such that they only promoted growth in the absence of arabinose at 30 °C, but not 37 °C or 42 °C. Using a linked yadC::Tn10 marker, four unique alleles of ponB were transduced to a fresh MM33(attλTB309) [ΔlpoB ΔponA (Para::ponA)] background, and the LpoB-bypass phenotype was confirmed (Fig. 2). Moreover, expression constructs producing GFP-PBP1b* variants specifically restored viability to PBP1a− LpoB− cells (SI Appendix, Fig. S1). We therefore conclude that amino acid substitutions E313D, I202F, Q411R, and Q447K in PBP1b are sufficient to bypass the LpoB requirement for PBP1b function in vivo. Variants with the substitutions E313D, I202F, and Q411R are characterized further here. The PBP1b(Q447K) variant was isolated in a separate selection after the completion of these downstream studies, and therefore was not subjected to additional analysis.
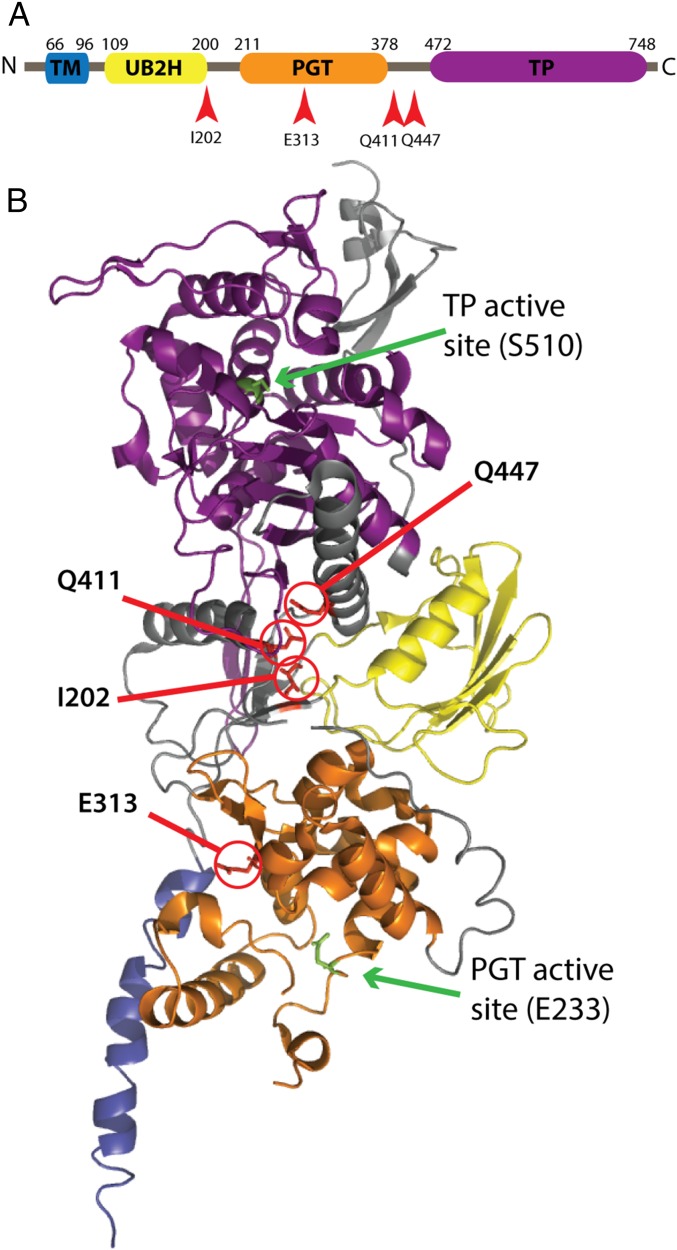
Fig. 1.
Location of altered residues in PBP1b* variants. (A) Schematic of the PBP1b primary structure: transmembrane helix (TM), UB2H domain, peptidoglycan glycosyltransferase (PGT), and transpeptidase (TP). (B) Crystal structure of PBP1b (PDB: 3FWM) (19) with the locations of the altered residues in the PBP1b* variants highlighted in red, along with the catalytic residues for both active sites in green. Domains in the structure are colored to match those in A.
Fig. 2.
Suppression of the PBP1a− LpoB− synthetic lethal phenotype by PBP1b* variants. Cells of TU121(attλTB309) [ΔponA (Para::ponA)], MM33(attλTB309) [ΔponA ΔlpoB (Para::ponA)], and the indicated MM33(attλTB309) derivatives [MM100, MM102, MM104, and MM105 all with (attλTB309)] harboring the indicated PBP1b* variant were grown overnight in M9 medium with 0.2% arabinose at 30 °C. The resulting cultures were normalized to an OD600 of 2.0 serially diluted, and 5 µL of each dilution was spotted onto the indicated medium. Plates were incubated overnight at 30 °C.
PBP1b* Variants Mimic LpoB Activation in Vitro.
Because overproduction of PBP1b(WT) does not suppress LpoB inactivation (9) (SI Appendix, Fig. S1), one of the simplest models to explain the LpoB-bypass phenotype of the PBP1b* variants is that the proteins have gained the ability to synthesize PG in the absence of LpoB. To investigate this possibility, we monitored PG synthesis in ether-permeabilized cells (EPCs) by supplying them with UDP-MurNAc-pep5 and radiolabeled UDP-[14C]GlcNAc (9). EPCs use the soluble precursors to produce lipid-II for PG synthesis by the PBPs. Importantly, PG synthesis in EPCs has been shown to depend principally on PBP1b function; EPCs prepared from ΔponB or ΔlpoB cells show greatly reduced synthesis (9) (Fig. 3A).
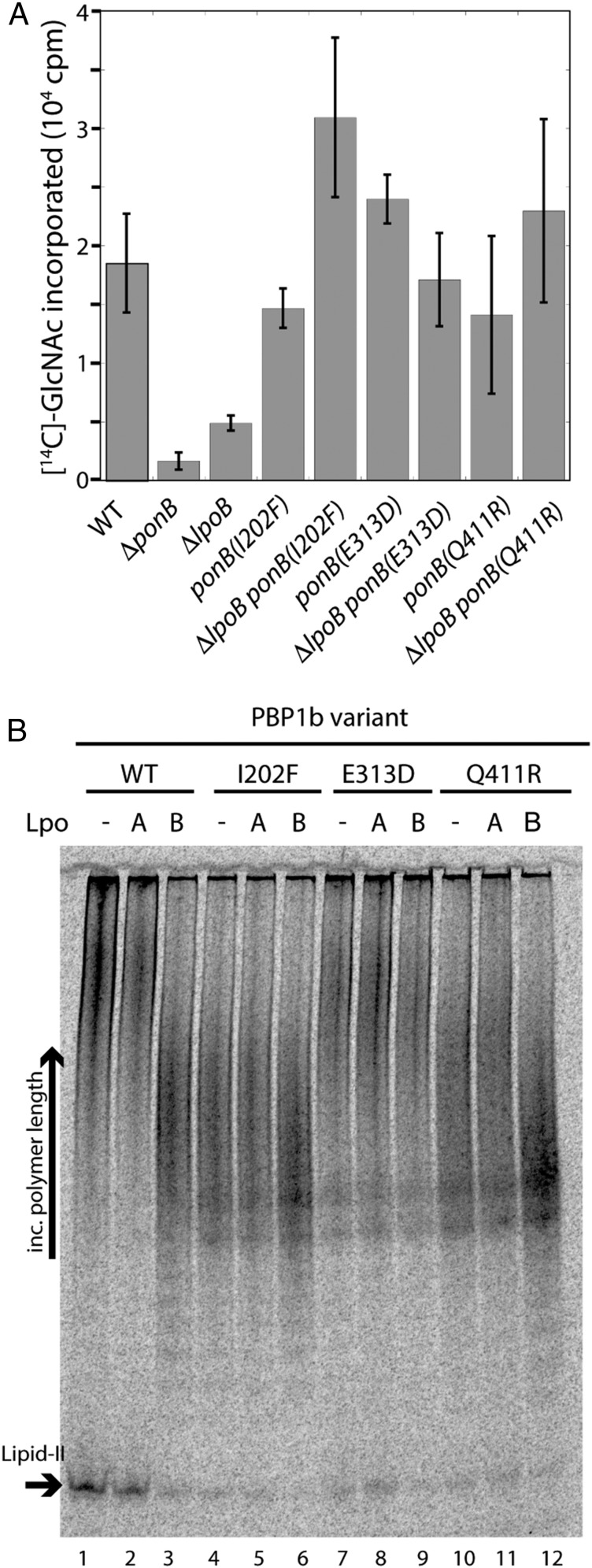
Fig. 3.
PBP1b* variants produce PG in the absence of LpoB. (A) EPCs were prepared from strains MM127 [yadC::Tn10] and the indicated derivatives MM141, MM135, MM133, MM143, MM43, MM49, MM131, and MM142. Reactions with 0.18 mg total protein were initiated by the addition of UDP-MurNAc-pentapeptide and UDP-[14C]-GlcNAc. After 60 min, the reactions were boiled in 4% SDS and filtered. Labeled PG retained on the filter was quantified by liquid scintillation counting. EPCs were prepared from three independent cultures for each strain. Average PG synthesis activities and SDs (bars) are shown. (B) Purified PBP1b(WT) (400 nM) and the indicated derivatives were incubated with or without purified LpoA or LpoB (400 nM). PGT activity was then initiated with the addition of [14C]GlcNAc-labeled lipid-II (4 μM). Glycan chain products and remaining substrate were separated on an acrylamide gel (9%).
EPCs were prepared from cells producing PBP1b* variants in the presence or absence of LpoB. As expected, ΔlpoB EPCs with wild-type PBP1b displayed greatly reduced PG synthesis activity relative to the corresponding LpoB+ cells (Fig. 3A). Conversely, EPCs with the PBP1b* variants PBP1b(I202F), PBP1b(E313D), or PBP1b(Q411R) showed PG synthesis activity comparable to the wild-type control cells in both LpoB+ and LpoB− backgrounds (Fig. 3A). Thus, the EPC results suggest that the PBP1b* variants no longer require LpoB for their PG synthase activity. To investigate this possibility further, we purified the above PBP1b* variants and monitored their PGT activity, using a synthetic lipid-II substrate. Wild-type PBP1b converts radiolabeled lipid-II into long glycan polymers, which can be visualized by polyacrylamide gel electrophoresis (Fig. 3B, lane 1). Note, however, that after a 15-min reaction, a significant amount of lipid-II substrate remains unpolymerized. As shown previously (9), the addition of LpoB to PBP1b(WT) not only stimulates its PGT activity, as indicated by the near-complete conversion of lipid-II to polymer, but also alters the profile of polymers produced (Fig. 3B, lane 3). The glycan chains synthesized in the presence of LpoB are much shorter than those synthesized in its absence (9) (Fig. 3B, compare lanes 1 and 3). This effect is specific for LpoB, as the addition of the noncognate Lpo factor, LpoA, does not significantly affect PBP1b activity (9) (Fig. 3B, lane 2). Strikingly, the PGT activity of the PBP1b* variants alone appeared to be as robust as LpoB-activated PBP1b(WT), as indicated by the amount of lipid-II converted to product (Fig. 3B, compare lanes 1, 3, 4, 7, and 10). Importantly, the profile of glycan chains produced by PBP1b(I202F) and PBP1b(Q411R) in the absence of LpoB closely matched the shorter polymers made by LpoB-activated PBP1b(WT). However, although it appeared to have elevated PGT activity, PBP1b(E313D) produced long glycan chains both in the presence and absence of LpoB. We conclude that the PBP1b* variants bypass the LpoB-requirement in vivo because their amino acid substitutions promote the adoption of an activated PBP1b conformation (see Discussion).
Conditional Growth of Cells Producing PBP1b* Variants in the Absence of LpoB and PBP1a.
To further characterize the in vivo function of the PBP1b* variants, we attempted to construct strains harboring the activated proteins, in which the genes for both LpoB and PBP1a were deleted without an inducible copy of ponA. We successfully deleted lpoB in PBP1a+ strains producing the PBP1b* variants. However, we were surprisingly unable to delete both lpoB and ponA in cells producing PBP1b(I202F) or PBP1b(Q411R), even though they supported growth of LpoB− PBP1A− cells harboring an uninduced Para::ponA construct (Fig. 2). This result suggests that these variants require a low level of residual PBP1a activity to promote survival in the absence of LpoB. In contrast, we were successful in generating strain MM119 [ponB(E313D) ΔponA ΔlpoB] harboring the PBP1b(E313D) variant without its activator or a source of PBP1a. This strain grew poorly on standard LB medium with 0.5% NaCl, but normal growth was restored when cells were grown in LB lacking added NaCl (LB0N) (SI Appendix, Fig. S2). Strikingly, growth of MM119 cells was completely blocked when they were shifted to LB with 1% NaCl, a condition in which WT cells and strains related to MM119 harboring only two of the three mutations [ponB(E313D), ΔponA, or ΔlpoB] grew robustly (SI Appendix, Fig. S2A). The growth phenotype was salt-specific and was not caused by the addition of similar concentrations of other osmolytes (proline, sucrose, or glycerol) to the medium. Visual inspection of MM119 cells after the shift to LB-1%NaCl revealed that the growth defect resulted from catastrophic cell lysis occurring about 45 min after the change in medium (SI Appendix, Fig. S2B). The time-lag required for lysis suggests the defect occurs in response to continued growth in the presence of 1% NaCl, rather than an immediate bursting in response to a change in the osmotic conditions. The salt-dependent growth of MM119 may reflect partial PBP1b(E313D) inactivation in LB-1%NaCl medium, such that it requires further stimulation by LpoB. Similarly, the failure of the PBP1b(I202F) or PBP1b(Q411R) variants to support growth without residual PBP1a when lpoB is deleted may also result from a partial LpoB dependence. Accordingly, the overproduction of GFP-PBP1b(E313D) or GFP-PBP1b(Q411R) allowed the construction and growth of ΔponA ΔlpoB strains even on LB-1%NaCl medium (SI Appendix, Fig. S3). This observation indicates that the main defect in the ΔponA ΔlpoB strains harboring the PBP1b* variants is limiting aPBP activity. Thus, although the PGT activity of the PBP1b* synthases appears to be maximally active and unresponsive to LpoB in vitro, the synthases retain some level of LpoB dependence in vivo (see Discussion).
A LpoB–PBP1b Complex Is Not Required to Bridge the Inner and Outer Membrane.
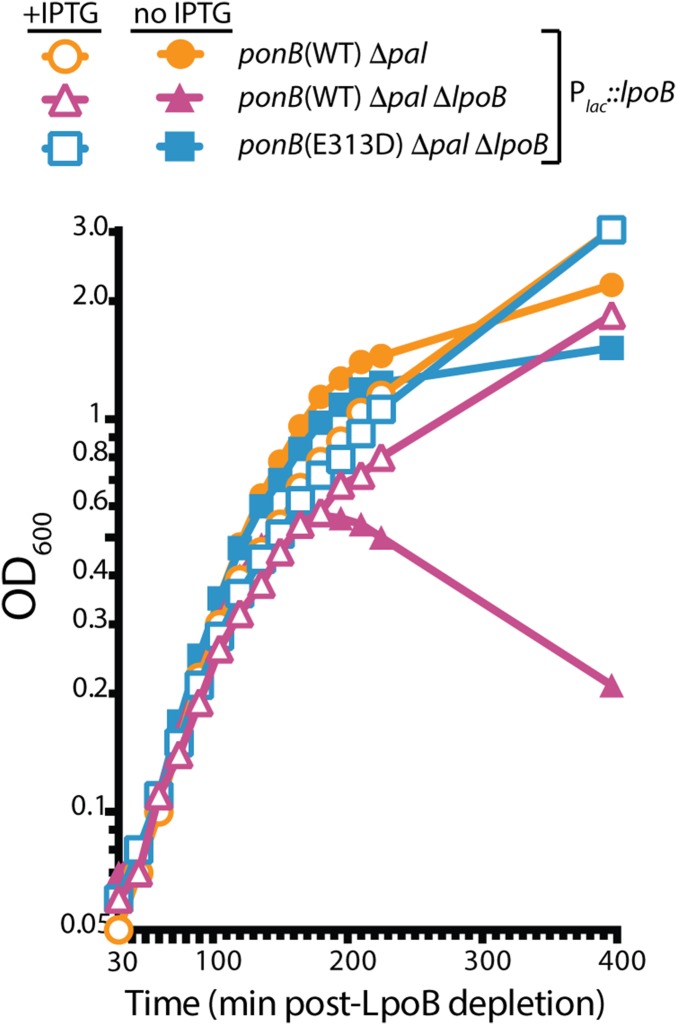
It was previously shown that defects in the Tol–Pal system result in severely impaired cell growth in LB0N when LpoB or PBP1b are also inactivated (10). The Tol–Pal system forms a transenvelope complex that is thought to mediate outer membrane constriction during cell division (17). Because of the observed negative genetic interaction between the Tol–Pal and PBP1b-LpoB systems, it was proposed that the PBP1b–LpoB complex provides an important physical link between the inner and outer membranes that is partially redundant with the connection provided by Tol–Pal (10). However, an alternative possibility is that it is a lack of PBP1b activity that causes the growth defect in LpoB mutants inactivated for Tol–Pal, rather than the loss of the outer membrane link. We used the LpoB-bypass mutants to test this model. Cells inactivated for the outer membrane lipoprotein Pal of the Tol–Pal complex grew to saturation in LB0N, whether or not additional LpoB is produced from a chromosomally integrated expression construct (Plac::lpoB) (Fig. 4). In contrast, and similar to prior observations (10), cells deleted for both pal and lpoB required induction of lpoB from the expression construct and lysed when LpoB was depleted (Fig. 4). The lysis phenotype of Δpal ΔlpoB cells depleted of LpoB was fully suppressed in cells producing PBP1b(E313D) (Fig. 4). Thus, cell lysis resulting from the inactivation of Tol–Pal in combination with LpoB likely results from a defect in PBP1b activity, rather than the loss of a redundant inner–outer membrane connection.
Fig. 4.
PBP1b(E313D) suppresses the growth defect of LpoB− Pal− mutants. Cells of MM109 [Δpal], MM115 [ponB(E313D) Δpal ΔlpoB], and MM117 [Δpal ΔlpoB], all containing the expression construct (attHKMM92) [Plac::lpoB], were grown overnight in LB supplemented with 250 µM isopropyl-β-D-thiogalactopyranoside (IPTG). The resulting cultures were diluted 1:100 and grown to an OD600 between 0.4 and 0.6 in the same medium at 37 °C. Cells were then washed three times in LB0N and inoculated into LB0N supplemented with either 0.2% glucose or IPTG (250 µM) to an OD600 of 0.025. Growth was then monitored at 37 °C.
Discussion
The aPBPs are complex enzymes capable of both polymerizing PG glycans and crosslinking them (1). Despite their status as important antibiotic targets, it remains largely unclear how their biochemical activities are controlled by cells to produce a uniform, cell-shaped PG matrix. An important step forward was the identification of outer membrane lipoprotein cofactors required for the in vivo activity of the E. coli aPBPs, PBP1a and PBP1b (9, 10). However, although these factors are known to stimulate aPBP activity in vitro, and their 3D structures have been solved (11–13, 18), very little is understood about the mechanisms by which the Lpo proteins promote PBP activity and whether the activation observed in vitro is physiologically relevant. Here, we addressed these issues by identifying a new class of PBP variants that bypass their normal cofactor requirement for in vivo function.
The amino acid changes identified in PBP1b, rendering it functional in the absence of LpoB, are located in two distinct regions of the structure. Three changes (I202F, Q411R, and Q447K) are clustered in the region between the major catalytic domains just behind the UB2H domain, where LpoB associates with PBP1b (10, 12) (Fig. 1). Variants with the I202F and Q411R substitutions were characterized biochemically and were shown to have elevated PGT activity. They also produced glycan chains with a length profile similar to the WT enzyme in the presence of LpoB, which are shorter than those produced by the WT enzyme alone. Thus, the PBP1b(I202F) and PBP1b(Q411R) variants behave in vitro as if they were bound by LpoB. The correlation of their elevated PGT activity in vitro and functionality in vivo in the absence of LpoB provides strong support for PGT activation being a physiologically relevant function of LpoB. In addition, the location of these substitutions near the UB2H domain suggests they induce a conformational change in PBP1b that mimics LpoB binding. Therefore, the results with these variants support a model in which LpoB controls PBP1b activity by stimulating its transition to a more active conformation, as opposed to interacting with the substrate to facilitate binding or other possible indirect modes of PBP1b activation. Further structural and biochemical characterization of these PBP1b variants should provide insight into the nature of the active conformation of the PBP1b PGT domain and how it is promoted by LpoB binding at the distantly located UB2H domain.
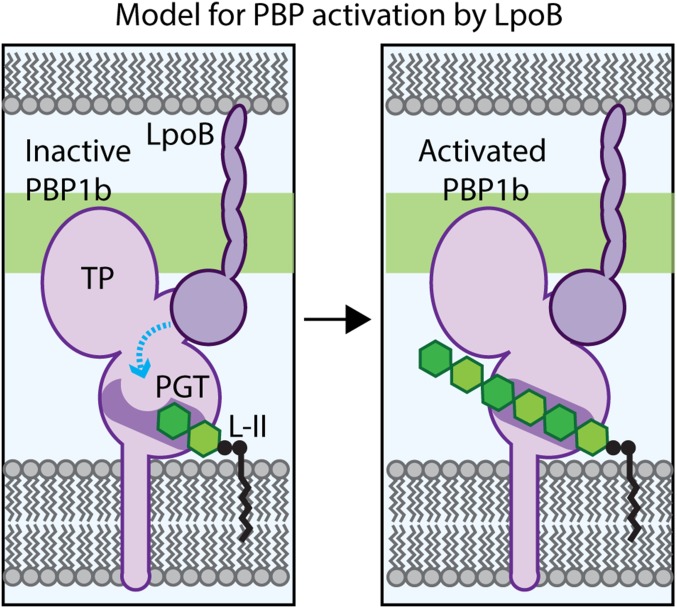
Unlike the other substitutions in PBP1b resulting in the LpoB-bypass phenotype, the E313D alteration is not proximal to the UB2H domain, but is instead located within the PGT domain. Similar to the I202F and Q411R changes, the E313D alteration also results in a stimulation of PBP1b PGT activity in vitro. However, the observed products more closely resembled the length distribution of glycans generated by the unstimulated WT enzyme than the shorter products produced by LpoB-activated PBP1b(WT) or the I202F and Q411R proteins, with or without LpoB. Moreover, the length of the glycans produced by PBP1b(E313D) was unaffected by LpoB addition. Residue E313 is located in the PGT domain in a cleft near where nascent glycan chains would be predicted to exit the polymerase (19). Shortening the side chain at this position by one methylene group (E to D) has a dramatic effect on enzyme activity, suggesting that E313 may be part of the mechanism that controls glycan polymerization. In the absence of LpoB, the side-chain of E313 may occlude glycan extension, and the proposed conformational change induced by LpoB may alter its location to remove the block and promote activity (Fig. 5).
Fig. 5.
Model for PBP1b regulation by LpoB. Shown is a schematic of the E. coli cell envelope with the inner membrane (Bottom) and partial outer membrane (Top) bracketing the PG layer (green). (Left) PBP1b is depicted in an inactive conformation, with the active site channel occluded in part by residue E313, such that the lipid-II (L-II) substrate cannot be elongated. We propose that the LpoB binding to the UB2H domain induces a conformational change in PBP1b (blue line) to alter the PGT active site, such that the channel opens (Right) to allow for lipid-II polymerization by the PGT domain and eventual crosslinking into the matrix by the TP domain.
An additional property of the E313D change is that it generates an enzyme that does not respond to LpoB to produce shorter glycans. We previously proposed that LpoB-activated PBP1b might produce shorter glycan polymers than PBP1b alone as a result of substrate competition (9). According to this scenario, most of the PBP1b molecules adopt an OFF conformation in the absence of LpoB. The few that are active, however, remain in the ON conformation and processively polymerize the supplied lipid-II to form long polymers. In the presence of LpoB, we proposed that a higher percentage of PBP1b molecules transition to the active conformation, therefore increasing competition for substrate such that it is exhausted before the enzymes can fully extend the polymers. The insensitivity of PBP1b(E313D) to the effect of LpoB on polymer length indicates that our previous substrate competition hypothesis is unlikely to be correct. Rather, the results suggest that LpoB and residue E313 not only control enzyme activity but also may also play direct roles in the poorly understood process of polymer length determination.
Part of the motivation for initiating this project was to understand the physiological function of LpoB. We suspected that cells producing constitutively active PBP1b variants might display a phenotype that helps explain why it is important for PBP1b activity to be controlled by LpoB in the first place. Despite testing numerous media types and growth temperatures, we did not observe a significant growth or morphological phenotype for PBP1b* producing cells with or without LpoB, as long as the PBP1a system was functional. The absence of a phenotype was surprising and suggests there may be additional levels of regulation operating to keep the activity of the PBP1b* variants in check. Further characterization of mutants producing these altered PBPs may reveal the nature of these controls.
In cells deleted for ponA, we were only able to delete lpoB in the ponB(E313D) background when cells were grown in LB0N. In higher-salt conditions (LB 1% NaCl), the triple ΔponA ΔlpoB ponB(E313D) mutant lysed and failed to form colonies. Similar attempts to generate mutants deleted for both ponA and lpoB in the ponB(I202F) and ponB(Q411R) backgrounds failed at all salt concentrations. In contrast, ponA can be deleted in all of the strains producing PBP1b* variants, regardless of NaCl concentration, when either LpoB is present or residual PBP1a is produced from an uninduced Para::ponA construct. Therefore, the fundamental defect of ΔponA ΔlpoB ponB* cells appears to be limiting aPBP activity. Consistent with this possibility, deletion of both ponA and lpoB was made possible at all NaCl concentrations when PBP1b(E313D) or PBP1b(Q411R) were overproduced. Overall, these observations suggest the PBP1b* variants retain the ability to be stimulated by LpoB in vivo, even though the enzymes alone appear to be as active as the LpoB-stimulated wild-type protein in vitro. Future work will be required to determine whether this remaining LpoB dependence reflects enhanced PGT stimulation by LpoB in vivo that is not detected in vitro, or whether LpoB possesses an additional activity unrelated to PGT activation responsible for promoting optimal PBP1b activity in vivo.
One possible alternative function previously proposed for LpoB is the formation of a bridge with PBP1b to connect the inner and outer membranes and facilitate outer membrane invagination during cell division (10). This proposal is based on the synthetic lethal phenotype of cells defective for both LpoB and the Tol–Pal system. Although possible, an alternative explanation for the negative genetic interaction is that PBP1b activity becomes especially important when the Tol–Pal system is defective. Our results support this latter explanation. PBP1b(E313D) suppresses the synthetic lethal interaction between LpoB and Pal without a bridge to the outer membrane. Thus, although recent results further connect the activities of PBP1b and the Tol–Pal system (20), the coordination is unlikely to involve a direct role for the PBP1b–LpoB complex in outer membrane constriction.
In this report, we used a combination of genetics and biochemistry to begin dissecting the molecular mechanisms underlying the control of cell wall biogenesis. Although the results focused on PBP1b and its control by the lipoprotein cofactor LpoB, the PBP1b variants identified here provide valuable reagents for future studies aimed at understanding the function of the aPBPs in general, and how the dual enzymatic activities of these important drug targets are regulated and coordinated.
Methods and Materials
Media, Bacterial Strains, and Plasmids.
Cells were grown in LB (1% tryptone, 0.5% yeast extract, 0.5% NaCl, unless otherwise indicated) or minimal M9 media supplemented with 0.2% Casamino Acids and 0.2% sugar, as indicated. All concentrations given as a percentage indicate wt/vol. The bacterial strains and plasmids used in this study are listed in the SI Appendix, Tables S2 and S3, respectively. All E. coli strains used in the reported experiments are derivatives of MG1655.
Selection and Screen for LpoB-Bypass Mutants.
Cells of MM33(attλTB309) [ΔlpoB ΔponA (Para::ponA)] were grown overnight in M9 medium containing 0.2% arabinose at 30 °C. The next morning, cultures were subcultured 1:100 into 50 mL of the same medium and grown to saturation at 30 °C. For the selection, 5- and 10-mL volumes of culture were pelleted, resuspended in 100 µL, and plated on M9 0.2% arabinose with 0.01 μg/mL cefsulodin. Resistant mutants arose at an average frequency of ∼10−7, based on two independent selections. Colonies were patched on M9 0.2% arabinose and M9 0.2% glucose agar to identify arabinose-independent isolates, which were considered good candidates for mutants encoding PBP1b* variants that function in the absence of LpoB. The ponB gene from each isolate was amplified and sequenced. All isolates harbored mutations in ponB. A list of the isolates and the mutations identified in the ponB gene are presented in SI Appendix, Table S1.
Protein Purification and Enzyme Assays.
PG synthesis in ether-permeabilized cells was monitored as described previously in ref. 9 with ether-treated cells prepared from cultures of the relevant strains grown at 30 °C. PBP1b (wild-type or variant) and Lpo proteins were purified as previously described (9). Glycan synthesis was assayed as described previously (9, 21). Wild-type PBP1b or PBP1b variants (400 nM) were preincubated with or without lipoproteins (400 nM) on ice for 30 min. Reactions were initiated on addition of proteins to lipid-II (4 µM) and incubated at room temperature for 15 min before heat inactivation (90 °C, 10 min). Gel electrophoresis analysis of the products was carried out as described previously (22).
Supplementary Material
Acknowledgments
We thank all members of the T.G.B., S.W., and D.E.K. laboratories for support and helpful comments. This work was supported by the National Institutes of Health (Grant R01AI083365 to T.G.B.; Grant AI099144 to T.G.B. and S.W.; Grant CETR U19 AI109764 to T.G.B., D.E.K., and S.W.; Grant GM76710 to D.E.K. and S.W.; and Grant GM066174 to D.E.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1524538113/-/DCSupplemental.
References
- 1.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10(2):123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruiz N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc Natl Acad Sci USA. 2008;105(40):15553–15557. doi: 10.1073/pnas.0808352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sham L-T, et al. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science. 2014;345(6193):220–222. doi: 10.1126/science.1254522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Müller P, et al. The essential cell division protein FtsN interacts with the murein (peptidoglycan) synthase PBP1B in Escherichia coli. J Biol Chem. 2007;282(50):36394–36402. doi: 10.1074/jbc.M706390200. [DOI] [PubMed] [Google Scholar]
- 5.Bertsche U, et al. Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol Microbiol. 2006;61(3):675–690. doi: 10.1111/j.1365-2958.2006.05280.x. [DOI] [PubMed] [Google Scholar]
- 6.Banzhaf M, et al. Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol Microbiol. 2012;85(1):179–194. doi: 10.1111/j.1365-2958.2012.08103.x. [DOI] [PubMed] [Google Scholar]
- 7.Kato J, Suzuki H, Hirota Y. Dispensability of either penicillin-binding protein-1a or -1b involved in the essential process for cell elongation in Escherichia coli. Mol Gen Genet. 1985;200(2):272–277. doi: 10.1007/BF00425435. [DOI] [PubMed] [Google Scholar]
- 8.Yousif SY, Broome-Smith JK, Spratt BG. Lysis of Escherichia coli by beta-lactam antibiotics: Deletion analysis of the role of penicillin-binding proteins 1A and 1B. J Gen Microbiol. 1985;131(10):2839–2845. doi: 10.1099/00221287-131-10-2839. [DOI] [PubMed] [Google Scholar]
- 9.Paradis-Bleau C, et al. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell. 2010;143(7):1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Typas A, et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 2010;143(7):1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jean NL, et al. Elongated structure of the outer-membrane activator of peptidoglycan synthesis LpoA: Implications for PBP1A stimulation. Structure. 2014;22(7):1047–1054. doi: 10.1016/j.str.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egan AJF, et al. Outer-membrane lipoprotein LpoB spans the periplasm to stimulate the peptidoglycan synthase PBP1B. Proc Natl Acad Sci USA. 2014;111(22):8197–8202. doi: 10.1073/pnas.1400376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King DT, Lameignere E, Strynadka NCJ. Structural insights into the lipoprotein outer membrane regulator of penicillin-binding protein 1B. J Biol Chem. 2014;289(27):19245–19253. doi: 10.1074/jbc.M114.565879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lupoli TJ, et al. Lipoprotein activators stimulate Escherichia coli penicillin-binding proteins by different mechanisms. J Am Chem Soc. 2014;136(1):52–55. doi: 10.1021/ja410813j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curtis NA, Orr D, Ross GW, Boulton MG. Affinities of penicillins and cephalosporins for the penicillin-binding proteins of Escherichia coli K-12 and their antibacterial activity. Antimicrob Agents Chemother. 1979;16(5):533–539. doi: 10.1128/aac.16.5.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singer M, et al. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53(1):1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PAJ. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol. 2007;63(4):1008–1025. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vijayalakshmi J, Akerley BJ, Saper MA. Structure of YraM, a protein essential for growth of Haemophilus influenzae. Proteins. 2008;73(1):204–217. doi: 10.1002/prot.22033. [DOI] [PubMed] [Google Scholar]
- 19.Sung M-T, et al. Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli. Proc Natl Acad Sci USA. 2009;106(22):8824–8829. doi: 10.1073/pnas.0904030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray AN, et al. Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. eLife. 2015;4 doi: 10.7554/eLife.07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye XY, et al. Better substrates for bacterial transglycosylases. J Am Chem Soc. 2001;123(13):3155–3156. doi: 10.1021/ja010028q. [DOI] [PubMed] [Google Scholar]
- 22.Barrett D, et al. Analysis of glycan polymers produced by peptidoglycan glycosyltransferases. J Biol Chem. 2007;282(44):31964–31971. doi: 10.1074/jbc.M705440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.