Fig. 5.
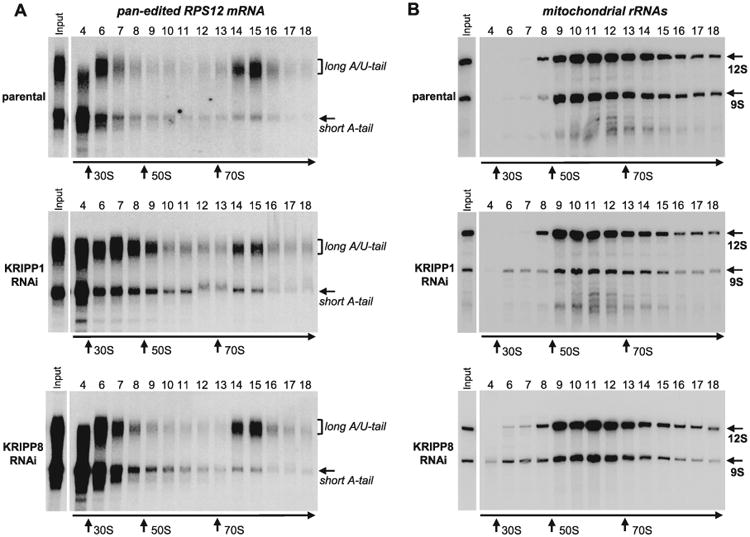
Sedimentation analysis of RPS12 mRNA and ribosomal RNAs in KRIPP1 and KRIPP8 knockdown cell lines. Detergent-extracted cell lysate from approximately 2 × 108 parasites was separated on 10–30% glycerol gradient. RNA was extracted from each fraction and separated in 5% polyacrylamide/8M urea gels for Northern blotting of mRNAs and rRNAs. All three membranes were hybridized simultaneously with probes specific for fully-edited RPS12 mRNA (A) and mitochondrial rRNAs (B). Purified bacterial ribosomal subunits and ribosomes were separated in parallel experiments as sedimentation standards. Arrows indicate peaks of optical density for apparent S value markers.
