Fig. 6.
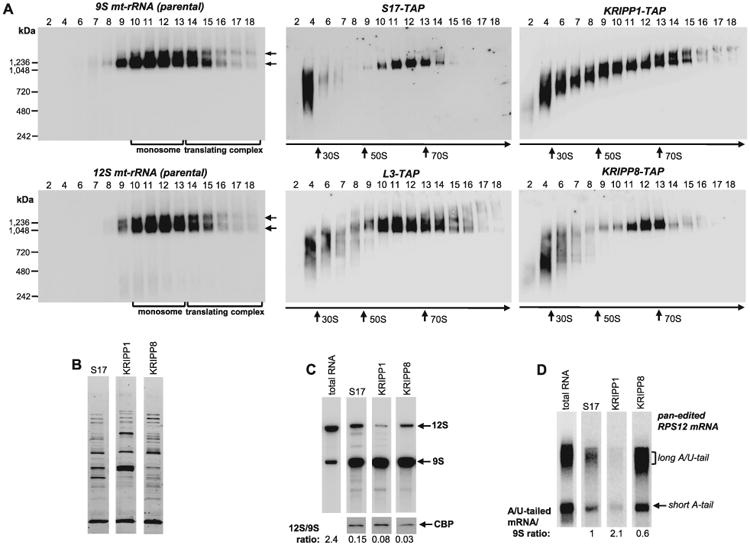
Distribution of KRIPP1 and KRIPP8 between ribosomal subunits, monosomes and mRNA-bound ribosomes.
A. Total cell lysates obtained from procyclic cell lines expressing indicated TAP-tagged fusions proteins were fractioned on glycerol gradients and each fraction was further separated on 3–12% Bis-Tris NuPAGE native gel. Native gels were transferred to nylon membrane, subjected to UV-crosslinking and hybridized with oligonucleotide probes specific for 9S and 12S mt-rRNAs (left panels). In parallel experiment, TAP-tagged proteins were detected by Western blotting with PAP reagent (central and right panels). NativeMARK unstained protein standards (Invitrogen) were separated on native gel alongside gradient fractions and stained separately. Sedimentation patterns of monosomes and translating complexes are indicated by arrows.
B. S17, KRIPP1 and KRIPP8 complexes were purified by rapid pulldowns, separated by SDS PAGE and stained with Sypro Ruby.
C. RNA was extracted from the parental cell line and from affinity purified complexes depicted in panel (B). RNA was separated on 5% polyacrylamide/8M urea gels for Northern blotting of mt-rRNAs (C) and edited RPS12 mRNA.
D. RNA amounts were normalized by protein bait in respective purified fractions, as quantitated by Western blotting with antibody against calmodulin binding peptide affinity tag. The 12S/9S ratio was calculated in relative intensity units by hybridizing the membrane with both probes simultaneously. The A/U-tailed mRNA ratio to 9S mt-rRNA was calculated from relative intensities of A/U-forms assuming such value in S17 preparation as 1.
