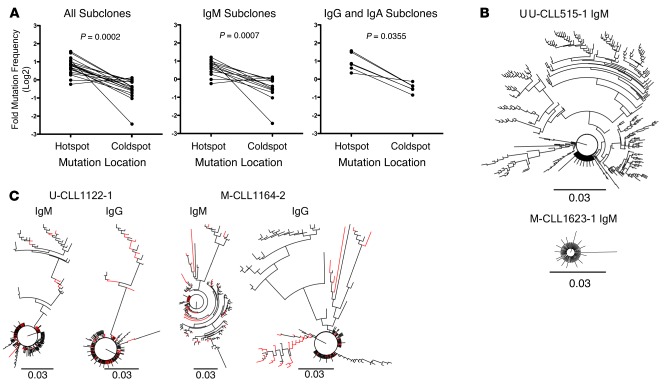
Figure 4. Xenografted chronic lymphocytic leukemia (CLL) B cells demonstrate clonally related IgM and IgG IGHV-D-J mutations that exhibit hallmarks of AID action.
(A) AID hotspot and coldspot mutation frequencies were calculated from new IGHV-D-J mutations in 19 xenografted samples and log2 transformed for statistical analysis by Wilcoxon test (P = 0.0002, left). This indicated that AID hotspot mutation frequencies are significantly higher. The same was observed among IgM subclones (n = 13; P = 0.0007, center) and among Ig isotype-switched subclones (n = 6; P = 0.0355, right). (B) Representative phylogenetic relationships of new subclones are illustrated using a polar tree layout, with each branch tip representing a distinct subclone. The length that each branch extends from the circle is roughly proportional to the number of mutations (K80 phylogenetic distance, scale bar: 0.03). Subclones with single mutations are indicated by branch tips extending a short distance from the circle, while multiple-mutated subclones are represented by tips extending farther out. The branches connecting each tip illustrate the sequence-relatedness between subclones, with closely related sequences clustered together. Total number of subclones, defined by changes in DNA sequence from that of the initial clone and from those of subclones present in vivo at the time of sampling can be determined by counting the number of terminal branch tips at various lengths from the circle’s center. U-CLL515-1 IgM subclone IGHV-D-J sequences describe a tree with large multibranched relationships (386 subclones with 1–25 mutations). M-CLL1623-1 IgM subclone IGHV-D-J sequences define a tree with few branched relationships (91 subclones with 1–7 mutations).(C) Comparison of IgM and IgG phylogenetic relationships from same sample are shown as in B. IgM and IgG relationships are shown for U-CLL1122-1 (173 subclones with 1–26 mutations and 225 subclones with 1–28 mutations, respectively) and M-CLL1164-2 (135 subclones with 1–35 mutations and 144 subclones with 1–32 mutations, respectively) with shared sequences indicated as red branches. U-CLL, CLL clone with IGHV sequence differing ≤2% from most similar germline gene; M-CLL, CLL clone with IGHV sequence differing >2% from most similar germline gene.