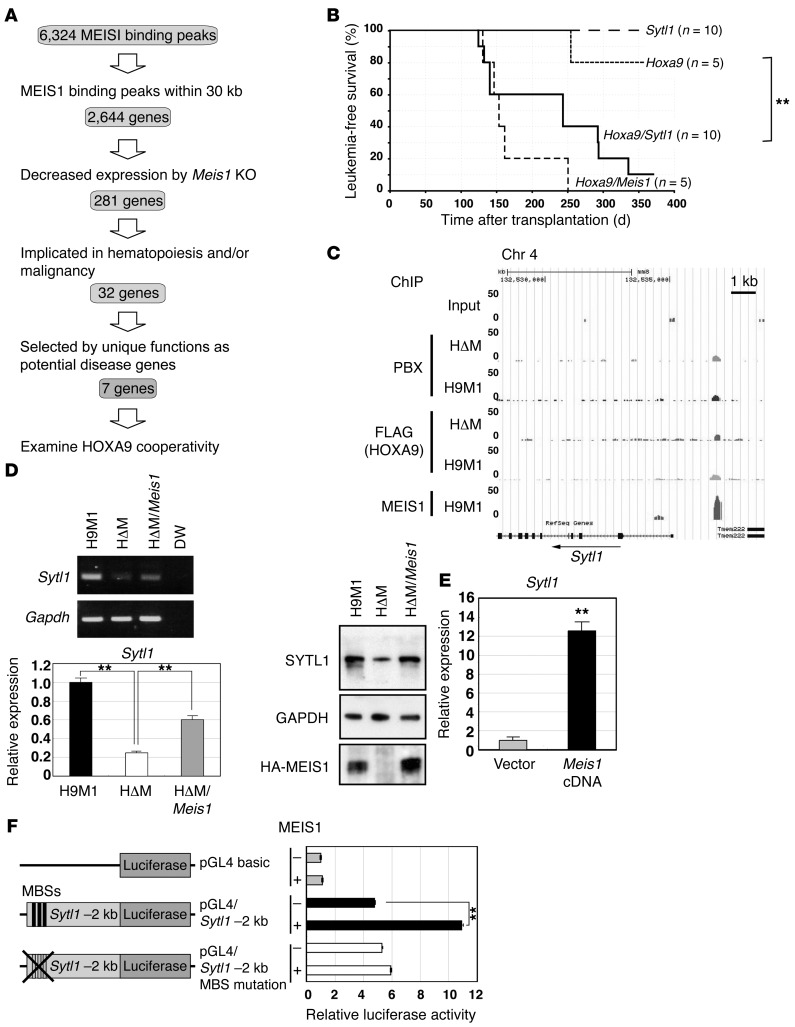
Figure 3. Sytl1 is a MEIS1 transcriptional target in leukemogenesis.
(A) Schematic diagram for target gene identification. (B) Sytl1 cooperated with Hoxa9 in leukemogenesis. Kaplan-Meier survival curves for recipient mice transplanted with Hoxa9-, Sytl1-, Hoxa9/Sytl1-, or Hoxa9/Meis1-transduced bone marrow cells. **P = 0.015, log-rank test. (C) ChIP-Seq analysis showed a MEIS1-binding peak 2 kb upstream of Sytl1. (D) Downregulation of Sytl1 by Meis1 KO in H9M1 cells. Representative gels of RT-PCR (3 experiments) and Q-RT-PCR (3 experiments) for Sytl1 mRNA expression and immunoblotting for the SYTL1 protein using α-SYTL1 in H9M1 (3 experiments). Immunoblotting for GAPDH and HA-MEIS1 proteins was performed on separate gels. Mean values of relative Sytl1 mRNA expression were normalized to Gapdh mRNA (**P < 0.01, 1-way ANOVA with Dunnett’s multiple comparison test). (E) Sytl1 expression was upregulated by Meis1 overexpression in 32Dcl3 cells. Sytl1 mRNA expression relative to Gapdh mRNA based upon Q-RT-PCR (n = 3, **P < 0.01, 2-tailed Student’s t test). (F) Luciferase reporter assay. A 2-kb fragment upstream of Sytl1 was introduced upstream of the luciferase cDNA. The mutant clone was created by introducing TGACAG to TGTTAG mutations into 3 putative MEIS1-binding sequences (MBS). Relative luciferase activities are shown in the presence or absence of MEIS1. Data represent mean ± SEM of 3 independent experiments (**P < 0.01, 2-tailed Student’s t test).