FIGURE 4.
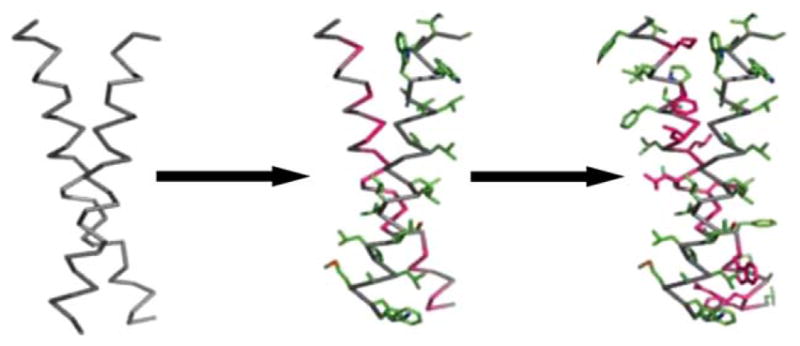
The CHAMP design process. A transmembrane helix-helix backbone pair with the desired geometry is selected from a database of membrane-protein structures (left). The original amino acid side-chains are discarded, and the helical backbones are extended to span the full length of a membrane. Next, the sequence from the target TM protein is “threaded” onto either one of the helices—in this example, the right helix with green side-chains (middle). Last, the sequence and side-chain configurations for the anti-TM peptide (represented by the left helix) is chosen via iteration over amino acids and rotamer re-packing (right). From Yin, H.; Slusky, J. S.; Berger, B. W.; Walters, R. S.; Vilaire, G; Litvinov, R.I.; Lear, J. D.; Caputo, G. A.; Bennett, J. S.; DeGrado, W. F. Science 2007, 315(5820), 1817–1822. Reprinted with permission from AAAS.
