Figure 4.
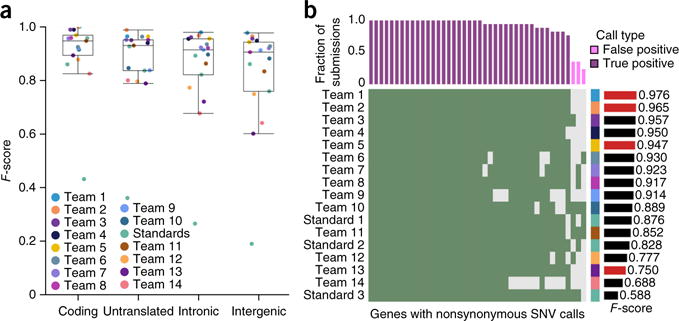
Effects of genomic localization. (a) Box plots show the median (line), interquartile range (IQR; box) and ±1.5× IQR (whiskers). For IS1, F-scores were highest in coding and untranslated regions and lowest in introns and intergenic (P = 6.61 × 10−7; Friedman rank-sum test). (b) Rows show individual submissions to IS1; columns show genes with nonsynonymous SNV calls. Green shading means a call was made. The upper bar plot indicates the fraction of submissions agreeing on these calls, and the color indicates whether these are FPs or TPs. The bar plot on the right gives the F-score of the submission over the whole genome. The right-hand side covariate shows the submitting team. All TPs are shown, along with a subset of FPs.
