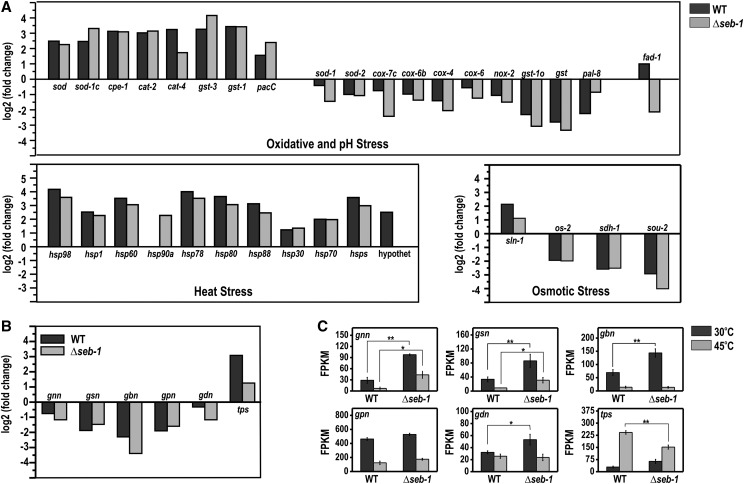
Figure 10.
SEB-1 controls the expression of genes involved in glycogen and trehalose metabolism and some stress responsive genes under heat stress. The expression levels were indicated as fold change (log2) and FPKMs (fragments per kb of transcript per million mapped reads). (A) Genes involved in oxidative, pH, heat, and osmotic stress responses in wild-type (WT) and Δseb-1 strains exposed to heat stress (45°). (B) Genes involved in glycogen and trehalose metabolism in WT and Δseb-1 strains exposed to heat stress (45°). (C) Genes involved in glycogen and trehalose metabolism in the WT and Δseb-1 strains exposed or not exposed to heat stress (45°). gnn, glycogenin (NCU06698); gsn, glycogen synthase gene (NCU06687); gbn, glycogen branching enzyme gene (NCU05429); gpn, glycogen phosphorylase gene (NCU07027); gdn, glycogen debranching enzyme gene (NCU00743); sod, superoxide dismutase variant gene (NCU09560); sod-1, superoxide dismutase-1 gene (NCU02133); sod-2, superoxide dismutase-2 gene (NCU01213); cat-2, catalase-2 gene (NCU05770); cat-4, catalase-4 gene (NCU05169); os-2, osmotic sensitive-2 gene (NCU07024); pacC, pH-response transcription factor pacC/RIM101 gene (NCU00090). The error bars represent the standard deviation for each condition, with triplicate measurements. Values of three replicates were used for statistical analysis and the significances (*P < 0.01 and **P < 0.001) between strains were estimated by the Tukey-Kramer multiple comparison test.