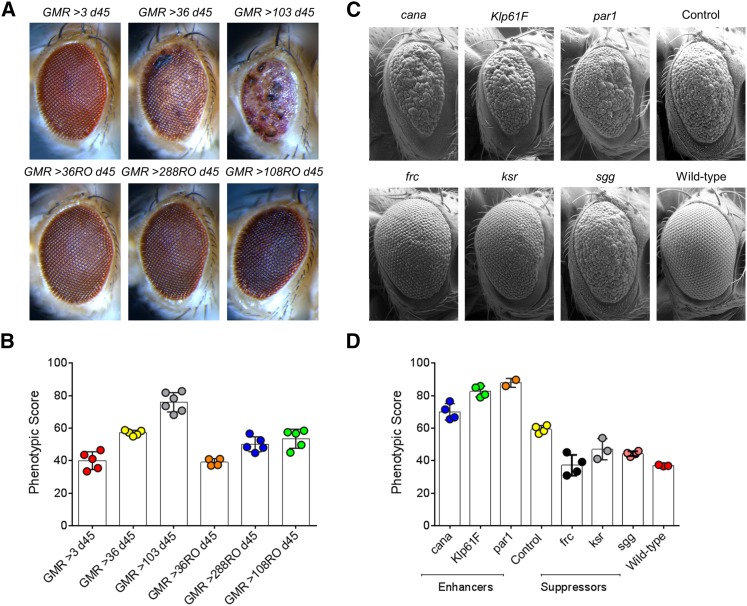
Figure 5.
Analysis of eye images obtained from independent studies. (A) Bright-field microscopy images of representative Drosophila eyes overexpressing C9orf72 pure or RO repeats using the GMR-GAL4 driver, imaged on d 45. While three pure repeats had no effect, 36 pure repeats were toxic and 103 pure repeats showed more overt toxicity; 36 RO repeats had no effect, and 108 RO and 288 RO repeats showed a mild effect. (B) Graph representing the phenotypic scores of C9orf72 pure or RO repeats using the GMR-GAL4 driver. The phenotypic scores are concordant with the visual assessment of the eye phenotypes (Mizielinska et al. 2014). The numbers of images used for these assays were n = 4 for GMR >36 RO d45, n = 5 each for GMR > 3 d45, GMR > 36 d45, GMR > 288 RO d45, and GMR > 108 RO d45, and n = 6 for GMR > 103 d45. (C) Scanning electron microscope images of genetic modifiers of tau-induced neurotoxicity. The control listed is w1118/+;gl-tau/+. All other panels, except wild type, contain one copy of gl-tau transgene in trans to one disrupted copy of the gene listed in the panel. (D) A graph representing the phenotypic scores of wild type, control, and the three enhancers and suppressors of w1118/+;gl-tau/+, is shown. The number of images used for these analyses were n = 2 for par1, n = 3 each for wild type, and ksr, n = 4 each for control, cana, Klp61F, sgg, and frc.