Figure 1. Strategy to identify silkworm Trimmer.
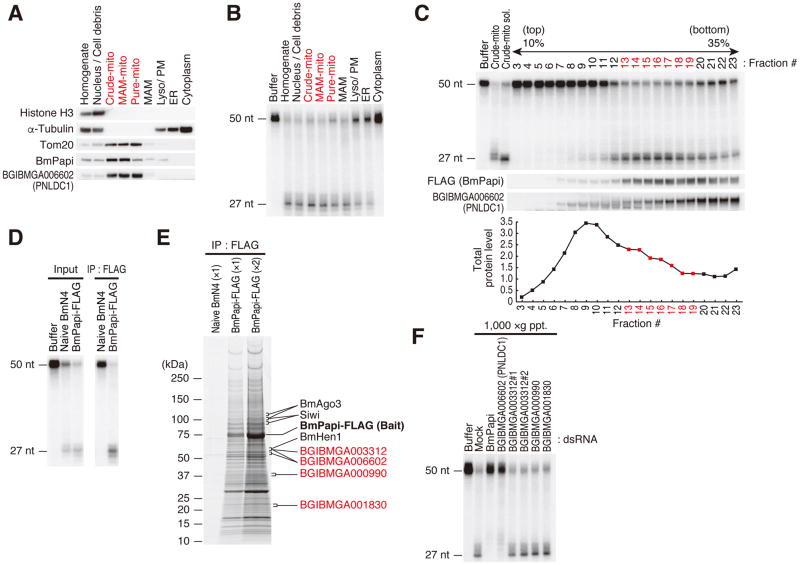
(A) Western blot analysis of subcellular fractionated BmN4 cells. Equal amounts of total protein in each fraction were loaded. Histone H3, α-Tubulin and Tom20 were used as fraction markers of nucleus, cytoplasm and mitochondria, respectively. The crude mitochondrial (crude-mito) fraction was further separated into MAM-mito, Pure-mito, and MAM fractions. MAM, mitochondria associated membrane; Lyso/PM, lysosome and plasma membrane; ER, endoplasmic reticulum. Mitochondria enriched fractions were indicated in red. BmPapi and Trimmer are enriched in the mitochondria-containing fractions.
(B) In vitro trimming assay using fractionated BmN4 cell lysate with an equal protein concentration. The mitochondria-containing fractions showed high trimming activity.
(C) CHAPS-solubilized crude mitochondrial (crude-mito) fraction prepared from BmPapi-FLAG stable cells was separated by 10–35% sucrose density gradient centrifugation. In vitro trimming assay was performed using each fraction (top panel). The distribution of BmPapi and Trimmer was analyzed by Western blotting (middle panel). Total protein level in each fraction is shown (bottom panel). The distribution of BmPapi and Trimmer correlated well with the trimming activity. The fractions with high trimming activity are indicated in red.
(D) CHAPS-solubilized crude mitochondrial (crude-mito) fractions were prepared from naive BmN4 or BmPapi-FLAG stable cells and subjected to immunoprecipitation with anti-FLAG antibody. In vitro trimming assay was performed using CHAPS-solubilized crude-mito fraction (IP input, left panel) or FLAG immunoprecipitates (right panel). The BmPapi-FLAG complex showed clear trimming activity.
(E) The immunopurified BmPapi-FLAG complex was resolved by SDS-PAGE and stained with Oriole fluorescent gel stain reagent. Four 3′–5′ exonucleases identified by mass spectrometry in the BmPapi-FLAG complex are shown in red. Details are shown in Table S1.
(F) BmN4 cells were transfected with dsRNAs for Trimmer candidates. In vitro trimming assay was performed using 1,000 ×g pellet fractions. For BGIBMGA003312, two different dsRNAs were used (#1 and #2). Knockdown efficiency is shown in Figure S1C. Mock indicates BmN4 cells transfected with dsRNAs for Renilla luciferase. Only the knockdown of BGIBMGA006602 decreased the trimming activity similarly to BmPapi knockdown.
See also Figures S1, S3 and Table S1.