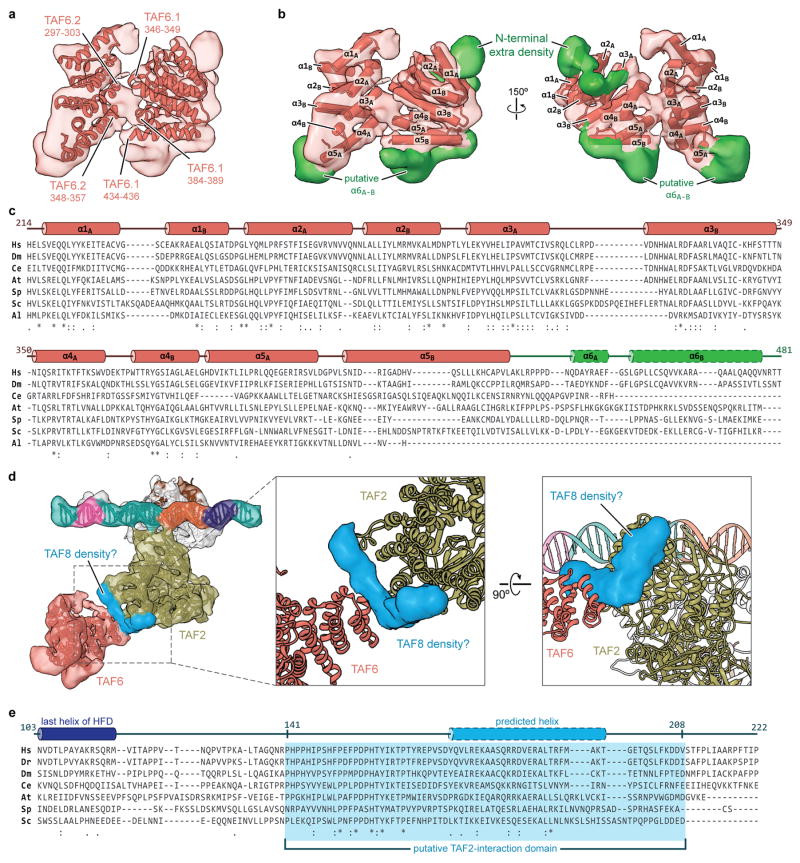
Extended Data Figure 6. Structural modeling and conservation of TAF6 and putative TAF8 density.
a, Cryo-EM density of the TAF6 dimer with fitted homology models. Putative regions involved in the homodimer interface are labeled. b, Organization of α-helices within the human TAF6 HEAT-like repeat and unaccounted density (green) around the TAF6 homodimer. c, Sequence alignment and secondary structure map of the TAF6 HEAT repeat domain (species abbreviations are the same as in Extended Data Fig. 4a, except that Al = A. locustae). The green region indicates the region that is unmodeled in our structure, with the two predicted C-terminal helices outlined with dashes. Numbering is based on the human sequence. d, Unaccounted density indicative of two α-helices, located between domain 4 of the TAF2 APD and one copy of the TAF6 HEAT domain, which we attribute to TAF8. e, Sequence alignment of a putative TAF2-interaction domain within TAF8 (species abbreviations are the same as in Extended Data Fig. 4a). The last helix of the structurally determined histone fold domain of TAF8 is depicted in dark blue, while the 26 residue stretch that is predicted to be α-helical is shown in light blue with dashed outline. Secondary structure prediction was performed with PSI-PRED71.