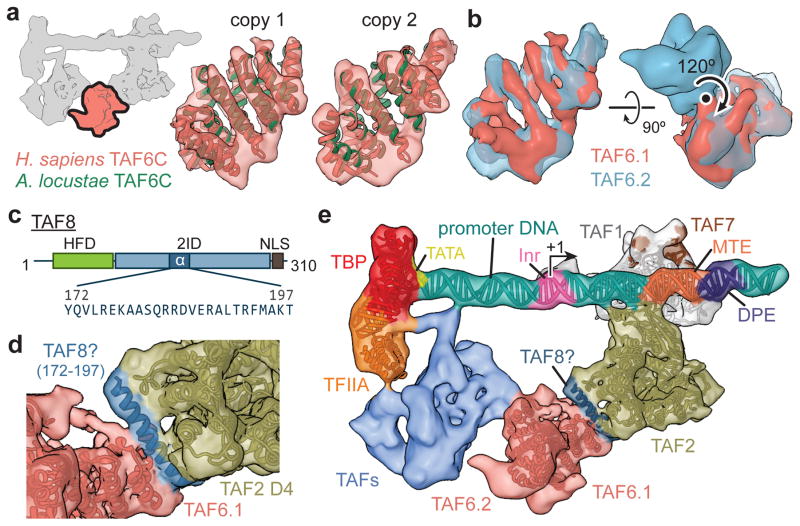
Figure 4. Structural TAFs within lobe C.
a, Docking of the crystal structure of A. locustae TAF6C (PDB code 4ATG)22 into two adjacent densities in the cryo-EM map, termed copy 1 and copy 2. The location of the segmented density in the overall map is highlighted in the schematic in the upper-left. b, The density for the two copies of TAF6C in the improved lobe C map are shown superimposed (left), and the homodimer interface and symmetry operation is depicted using the original map from Fig. 1b (right). c, Location and sequence of the predicted 26 residue helix within the TAF2-interacting domain (2ID) of TAF8. The relative locations of the histone fold domain (HFD) and nuclear localization signal (NLS) are also depicted. d, Docking of the TAF8 26 residue helix between TAF2 APD domain 4 (D4) and TAF6 copy 1 (TAF6.1). e, Overall architecture of TFIID with all fitted atomic models.