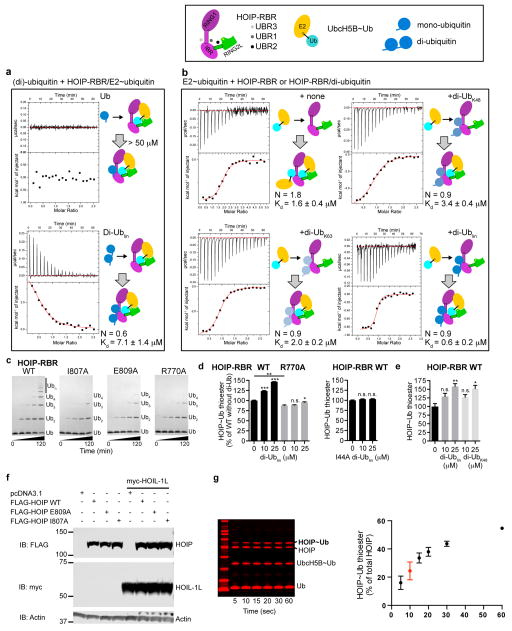
Extended Data Figure 9. UBR3 interacts with di-ubiquitin and allosterically promotes E2~ubiquitin binding and HOIP-RBR activation.
a, ITC experiments analyzing the binding of mono-ubiquitin or linear di-ubiquitin to an isolated HOIP-RBR/E2~ubiquitin complex. While the binding of mono-ubiquitin is below the sensitivity of the experimental setting (Kd > 50μM), the binding of linear di-ubiquitin exhibits a Kd of 7.1 μM. b, ITC experiments analyzing the binding of UbcH5B~ubiquitin to HOIP-RBR in the absence and presence of different di-ubiquitin chains. Top left: The binding stoichiometry of UbcH5B~ubiquitin and WT HOIP-RBR is n = 1.8, indicating that two UbcH5B~ubiquitin molecules interact with one RBR through UBR1/2 (catalytic binding) and UBR3 (binding of the ubiquitin moiety of the E2~ubiquitin conjugate) with a combined overall Kd of 1.6 μM. The graph shows a single titration step, indicating “crosstalk” between UBR3 and UBR1. Top right: The presence of K48-linked di-ubiquitin leads to 1:1 binding (n = 0.9) of UbcH5B~ubiquitin to HOIP-RBR, indicating that di-ubiquitin occupies UBR3 and limits UbcH5B~ubiquitin binding to only its bona fide catalytic binding site (with ubiquitin binding to UBR1/2 and UbcH5B binding to RING1/RING2L). However, the presence of K48-linked di-ubiquitin results in the lowest affinity (Kd = 3.4 μM) for the binding of the conjugate to the RBR, indicating a negative effect of this linkage compared to the other di-ubiquitin entities tested. Bottom left: K63-linked di-ubiquitin has a more favorable effect on UbcH5B~ubiquitin binding (Kd = 2.0 μM), Bottom right: The strongest allosteric effect is observed in the presence of linear di-ubiquitin, which enables sub-micromolar binding (Kd = 600 nM) of UbcH5B~ubiquitin. These results show that linear di-ubiquitin functions as a potent activator of HOIP-RBR by binding to UBR3 (see also below and Fig. 4). While the structure depicts the interactions of one ubiquitin unit with UBR3, a second ubiquitin C-terminal to the UBR3-interacting ubiquitin may undergo further interactions with the IBR (as indicated by the arrow in Extended Data Fig. 8c). The cartoon representations summarize the configuration of each ITC experiment. c, Polyubiquitination assays of UBR3 mutants. The HOIP-RBR I807A, E809A and R770A mutants exhibit a marked reduction in activity, supporting the importance of UBR3 in HOIP function. d, Activation of HOIP-RBR by di-ubiquitin. While wild-type HOIP-RBR is activated by the presence of increasing concentrations of WT linear di-ubiquitin, WT linear di-ubiquitin only has a weak effect on activation of the UBR3 R770A mutant (similar to the I807A and E809A mutants in Fig. 4b). Additionally the di-ubiquitin mutant I44A, which is mutated at a critical UBR3-interacting residue in both ubiquitin units, does not have an activating effect on WT HOIP-RBR thioester activity (mean activity ± s.e.m (n=3), one-way ANOVA followed by Tukey’s post-test, *: P<0.05, **P<0.01, ***: P<0.001, n.s.: not significant; representative gels shown in Supplementary Fig. 1). e, Effect of linear versus K48-linked di-ubiquitin on HOIP-RBR thioester transfer activity. In contrast to the ITC binding studies in panel b, linear and K48-linked di-ubiquitin are both able to increase the thioester transfer activity of HOIP-RBR (although the experimental setup necessary to investigate the K48-linkage resulted in larger error; see Methods; mean activity ± s.e.m (n=3), one-way ANOVA followed by Tukey’s post-test, *: P<0.05, **P<0.01, n.s.: not significant; representative gels shown in Supplementary Fig. 1). These results show an UBR3-dependent activating effect of di-ubiquitin, and thus potentially of polyubiquitin chains. However, whether HOIP-UBR3 acts as a universal ubiquitin sensor or has a preference for linear ubiquitin over other types of linkage needs to be further examined through careful investigations also including full-length proteins of the LUBAC in cellular settings. Additionally, although there is a substantial gap between UBR3 and the position of the acceptor ubiquitin, longer acceptor ubiquitin chains might be able to bridge this gap and mediate a cooperative effect between the two sites. This would be consistent with a recent publication showing that the presence of K63-linked ubiquitin chains is frequently necessary for the formation of linear polyubiquitin chains58. f, Protein expression levels for the NF-κB reporter assays in cells shown in Fig. 4d. Shown are anti-FLAG immunoblots of HOIP WT and mutants and anti-myc immunoblots of HOIL-1L, demonstrating similar protein expression levels in different cell lysates. Lysates were also probed by immunoblotting for actin as a loading control. Uncropped blots are shown in Supplementary Fig. 1. g, Time-course of HOIP~ubiquitin thioester transfer assay. (Left) SDS-PAGE showing time-course of HOIP~ubiquitin thioester transfer assay. Coomassie-stained bands in red visualized using LI-COR Odyssey at 700 nm. (Right) Plot of quantified HOIP~ubiquitin thioester transfer assay time-course (mean ± s.e.m., n = 2). The 10-second time point used in the end-point assays throughout the study is highlighted in red.