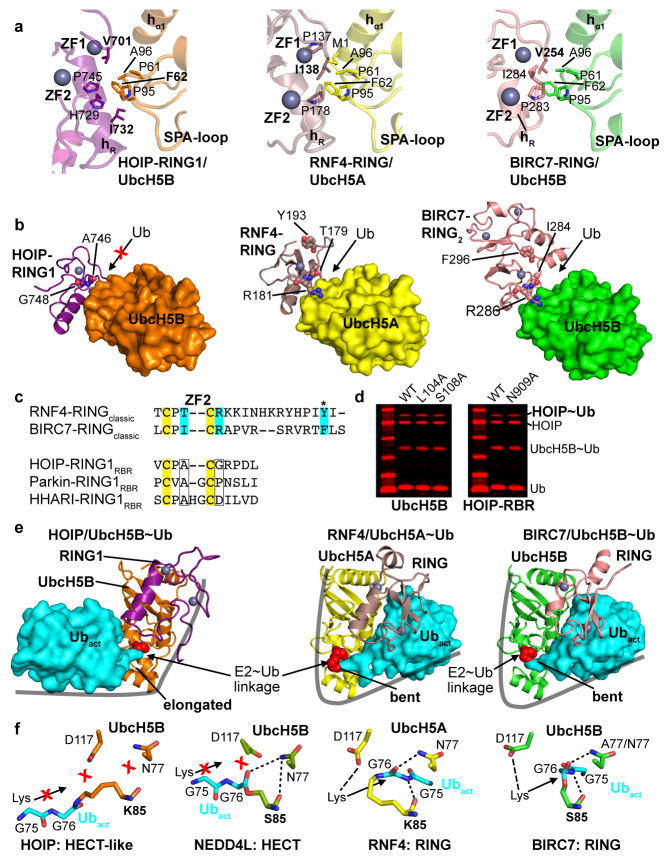
Extended Data Figure 4. The HOIP-RING1–IBR applies an altered binding mode compared to classic RING E3s necessitating a HECT-like mechanism.
a, The HOIP RBR-RING1 uses an E2 interaction pattern similar to classic RINGs, which however results in a shifted binding. Shown are the details of the RING/E2 interaction in the HOIP-RING1RBR/UbcH5B~ubiquitin complex (left), the RNF4-RINGclassic/UbcH5A~ubiquitin complex (middle; PDBid: 4AP421), and the BIRC7-RINGclassic/UbcH5B~ubiquitin complex (right; PDBid: 4AUQ24). The HOIP-RBR-type RING1 uses a pattern of hydrophobic residues as the core of the interaction with E2 that is similar to that in classic RING E3 ligases. Subtle differences however support a shifted binding mode (see also Fig. 2b). The main features of the RING and E2 as well as HOIP residues mutated in Fig. 2e – g are displayed in bold. Zinc finger (ZF) 1 and 2 of the RING domains and the SPA-loop of the E2 containing a conserved Ser-Pro-Ala motif are annotated. For the following panels the same structures and color codes as in panel a are used. b, The shift in binding and altered surface residues in HOIP-RING1 do not support the composite RING/E2 binding site for activated ubiquitin utilized by classic RING/E2 complexes. UbcH5A/B E2s are rendered as surface representation and the RING domains in ribbon representation. Residues crucial for classic RING E3s to recruit the activated ubiquitin in the composite RING/E2 ubiquitin-binding surface21,24 are depicted (middle, right). In HOIP-RING1 (left) equivalent residues are not conserved (displayed), indicating that HOIP cannot accommodate the activated ubiquitin in its RING1/UbcH5B complex. For illustration purposes, only one monomer of the dimeric RNF4 is shown (although Y193 from the other RING molecule is still displayed)21; for BIRC7 the RING dimer (RING2) is displayed24. c, Alignment of HOIP, Parkin and HHARI RING1 domains with classic RING domains centered around residues displayed in panel b. Residues crucial for ubiquitin binding in classic RINGs are highlighted in cyan and their structural equivalents in the RING1 domains of the RBR E3 ligases HOIP, Parkin and HHARI are indicated by boxes, attesting to the absence of a composite RING1/E2 ubiquitin binding site in RBR ligases (* indicates ubiquitin interaction residues from the other RING molecule in the dimers formed by the classic RINGs RNF4 and BIRC7). The T/I-C-R sequence observed in the dimeric RINGs of RNF4 and BIRC7 represents the highly conserved Φ-x-R/K motif, where Φ is a hydrophobic residue and x is either a Cys in RING E3 ligases or a polar residue in U-box ligases26. This motif is not only critical for E3-mediated catalysis by dimeric RING ligases (such as RNF4 or BIRC7) but is also necessary for E2-mediated catalysis by simpler monomeric RING and U-Box E3 ligases26. The fact that this motif is not conserved in HOIP, HHARI and Parkin further confirms the mechanistic differences between RBRs and classic RING domains. d. Thioester transfer assays show that E2 residues critical for classic RING-supported catalysis are not important for HOIP catalysis. Left: HOIP-RBR thioester transfer assays show similar activity of UbcH5B wild-type and L104A and S108A mutants. This is in stark contrast to the reported effects of these mutations on classic RING-supported catalysis21,24, underlying the fundamentally different mechanism of the HECT-like catalysis by HOIP-RBR. Right: Mutation to Ala of HOIP-RBR N909, which would be in the vicinity of the activated ubiquitin if the E2~ubiquitin conjugate were bound in a bent manner (see panel e), also shows no effect on HOIP thioester formation (Coomassie-stained bands in red). e, The altered E2~ubiquitin binding mode of RBR-RING1 results in the requirement for a HECT-like mechanism. Displayed are the entire RING/UbcH5~ubiquitin complexes with RINGs and E2s depicted in ribbon representation and the activated ubiquitin in surface representation. The bipartite binding mode used by the HOIP-RING1–hE2–IBR arm (see also Fig. 2a) results in an elongated E2~ubiquitin conformation (left, only the RING1 domain of HOIP is depicted) while formation of a composite RING/E2 binding surface in the case of classic RING E3 ligases (middle, right) results in binding of the activated ubiquitin in a compact manner with a bent E2~ubiquitin conformation. Importantly, this bent conformation places the thioester-link in a specific position relative to the catalytic machinery of the E2, allowing direct attack by the lysine/amine function of a substrate or growing ubiquitin chain. The Lys85/Ser85 residues mediating the E2~ubiquitin linkage and mimicking UbcH5A/B catalytic cysteine C85 are displayed as red spheres. In the elongated E2~ubiquitin conformation propagated by the HOIP-RBR, this attack is not possible. The linkage is however ideally positioned for the attack by the RBR catalytic cysteine in a HECT–like mechanism (see also Fig. 3 and Extended Data Fig. 7a). f, Close-up of the catalytic centers in E2~ubiquitin linkages. Details of the catalytic centers resulting from the E2~ubiquitin conjugate conformations outlined in panel e and, for comparison, the HECT-type E3 NEDD4L/UbcH5B~Ub structure (PDBid: 3JW028), with the directionality of an attacking amine indicated as previously proposed21. In the HECT-like RBR arrangement, the UbcH5B~ubiquitin linkage is not aligned correctly relative to the E2 catalytic machinery for a direct attack by an amine function. This is similar in the HECT-type arrangement in the NEDD4L complex but completely different from the arrangement in classic RING-supported E2 catalysis. Additionally, the ubiquitin C-terminal residues G75-G76 reside in a position that would overlap with the attacking amine. The available structure of the BIRC7/UbcH5B~Ub complex (PDBid: 4AUQ24) features an UbcH5B N77A mutant and the remainder of the Asn side-chain has been manually added based on wild-type UbcH5B from PDBid: 2ESK45 (right).