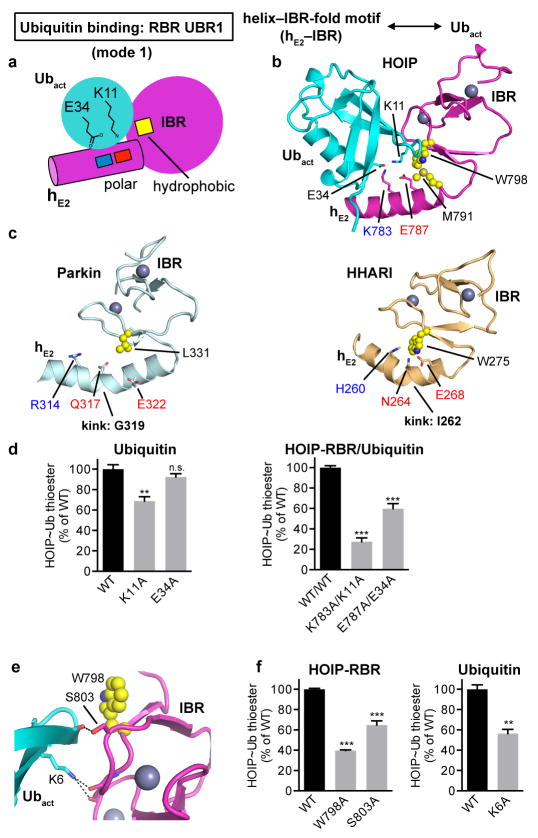
Extended Data Figure 5. Helix–IBR-fold motifs constitute new ubiquitin-binding regions (UBR) in active RBR proteins: Binding of the activated ubiquitin by UBR1 using binding mode 1.
a, Schematic illustrating binding mode 1 utilized by hE2–IBR to bind the activated ubiquitin in the RING1–IBR arm. The general principle of this binding mode is that the RING1 extension helix 2 (hE2) preceding the IBR presents a pattern of charged/polar residues (indicated by blue and red squares, which symbolize K, R, H and E, Q, N residues respectively) that interact with ubiquitin E34 and K11. These interactions are supported by the IBR surface, with a particular contribution of hydrophobic residues (yellow square) flanking the salt-bridge system. b, Coordination of the activated ubiquitin by HOIP-hE2–IBR in mode 1. HOIP-hE2 residues K783 and E787 bind ubiquitin residues E34 and K11 and are flanked by hydrophobic residues M791 and W798 from HOIP-IBR. c, Structurally equivalent residues in Parkin and HHARI. Displayed are the hE2–IBR modules from autoinhibited Parkin (PDBid: 5C1Z12) and HHARI (PDBid: 4KBL11) with residues equivalent to HOIP residues in panel b depicted, illustrating the general conservation of UBR1. It should be noted that these structures feature autoinhibited forms of the RBR proteins, which exhibit a kink in hE2 of UBR1. This kink would sterically hinder ubiquitin binding to UBR1 and likely participates in the RBR autoinhibition mechanism (see also Extended Data Fig. 8). d, Thioester-transfer assays of ubiquitin and UBR1 salt bridge mutants. In agreement with the observed four-residue salt bridge system in panel b, the single K11A or E34A ubiquitin mutations show only a slight to moderate effect since the remaining charged residue can still coordinate the two oppositely charged residues of HOIP. In contrast, elimination of both similarly charged residues in the complex by combining the HOIP K783A and ubiquitin K11A or HOIP E787A and ubiquitin E34A mutations results in a more dramatic loss of activity (mean activity ± s.e.m (n=3), one-way ANOVA followed by Tukey’s post-test, **P<0.01, ***: P<0.001, n.s.: not significant; representative gels shown in Supplementary Fig. 1). e, f, Role of the IBR in UBR1. Close-up of the additional IBR/Ubact interactions in stick representation (e) shows that HOIP S803 and ubiquitin K6 coordinate the backbone carbonyl functions of ubiquitin T12 and HOIP A800/K829 respectively. W798, which is involved in hydrophobic interactions, is also displayed in sphere representation. Quantitative thioester transfer assays (f) show that alanine mutants of residues outlined in panel e cause a significant loss of activity (mean activity ± s.e.m (n=3), left: one-way ANOVA followed by Tukey’s post-test, right: two-tailed unpaired Student’s t-test, **P<0.01, ***: P<0.001; representative gels shown in Supplementary Fig. 1).