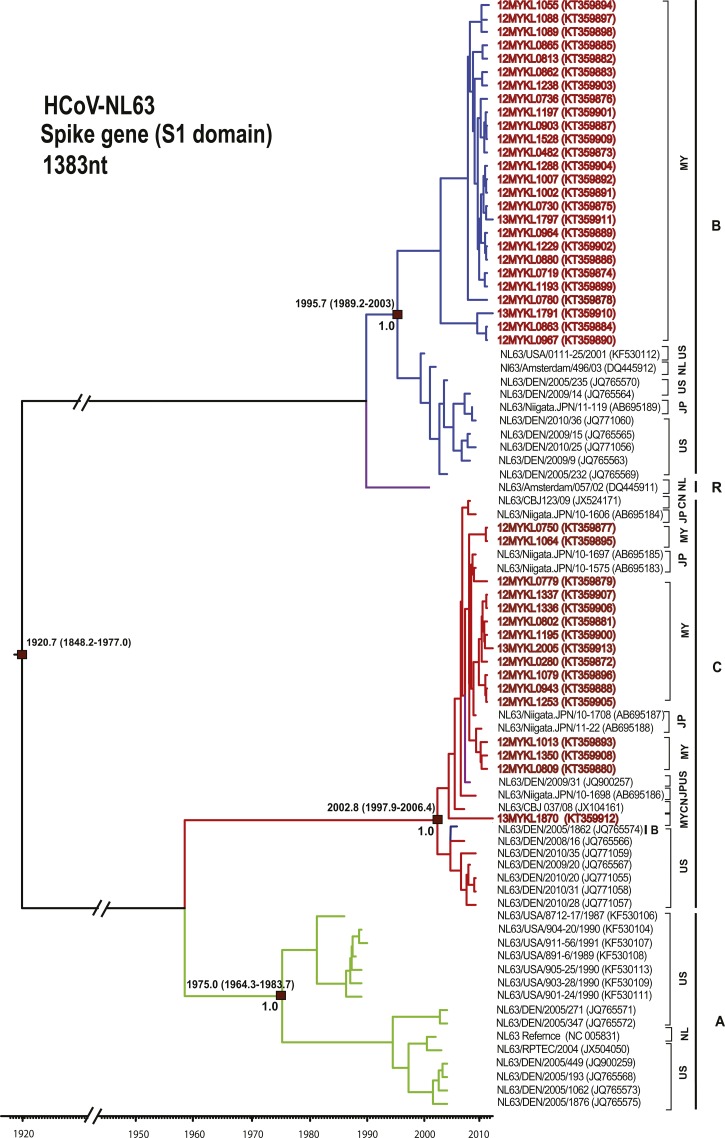
Figure 2.
Maximum clade credibility tree of HCoV-NL63. Spike gene (S1 domain) sequences (1,383 nucleotides) were analyzed under the relaxed molecular clock with a GTR+I substitution model and a constant size coalescent model implemented in BEAST. Posterior probability values and the estimation of the time of the most recent common ancestors with 95% highest posterior density were indicated on major nodes. The HCoV-NL63 sequences obtained in this study were color coded and HCoV-NL63 genotypes A–C were indicated; green = genotype A, blue = genotype B, and red = genotype C. The recombinant genotype is indicated by purple color. The sampling site for each sequence was indicated by codes for the representation of countries. Country codes are as follows; MY = Malaysia; US = United States; JP = Japan; NL = Netherlands; CN = China.