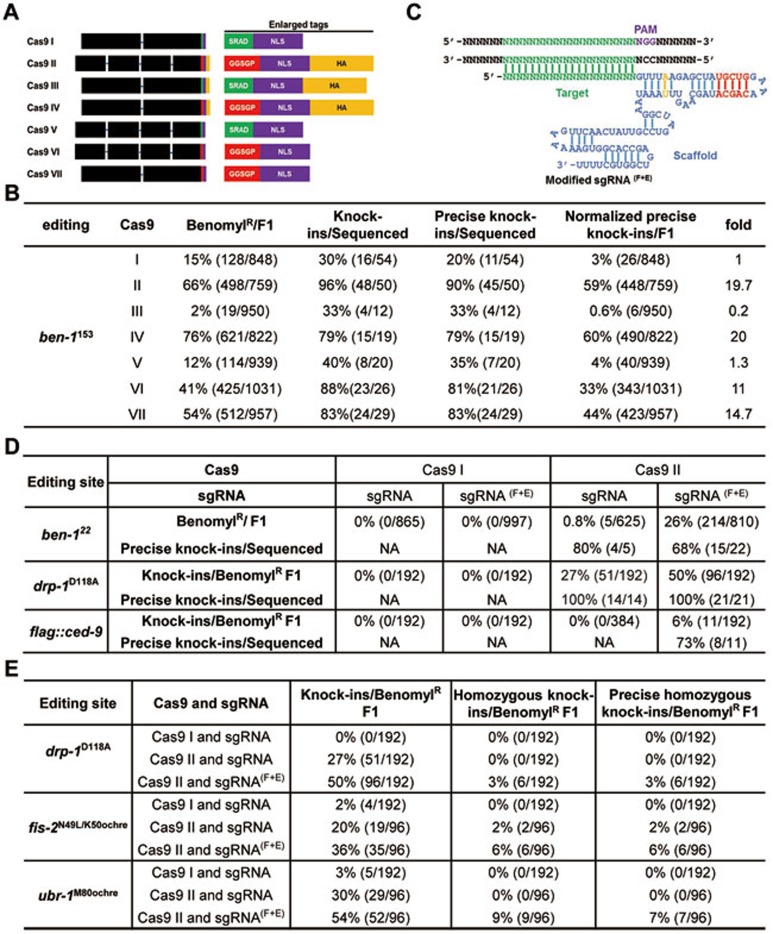
Figure 1.
A new combination of Cas9 and sgRNA greatly improves editing efficiency and fidelity and drives one-step homozygosity. (A) A schematic diagram of Cas9 proteins with different C-terminal tags. The black boxes depict the exons of the Cas9 coding region and the blue lines depict the introns. (B) The editing efficiencies at the ben-1153 position mediated by seven different Cas9 proteins shown in A. The normalized percentage of precise knock-ins identified from the total F1 animals screened is also shown. (C) A diagram of sgRNA(F+E) and its target DNA. The sequences shared by regular sgRNA and sgRNA(F+E) are in blue. An A-U base-pair flip and an extension of the first stem-loop in the scaffold of sgRNA(F+E) are highlighted in yellow and red, respectively. (D) Editing efficiencies of different combinations of Cas9 I or Cas9 II with regular sgRNA or sgRNA(F+E) at the indicated positions. For editing experiments at the ben-122 position, editing results were scored and presented as in B. For editing experiments at the drp-1118 position and the ced-9 locus, a co-CRISPR method, which includes a driver sgRNA proven to work well in editing, was used to facilitate the identification of edited F1 animals3,4. The ben-1153 sgRNA was used as a co-CRISPR driver. (E) Comparison of the editing efficiencies in generating F1 homozygotes with the indicated editings using different combinations of Cas9 I or Cas9 II with sgRNA or sgRNA(F+E). The co-CRISPR method was used to enrich F1 edited animals, as in D.