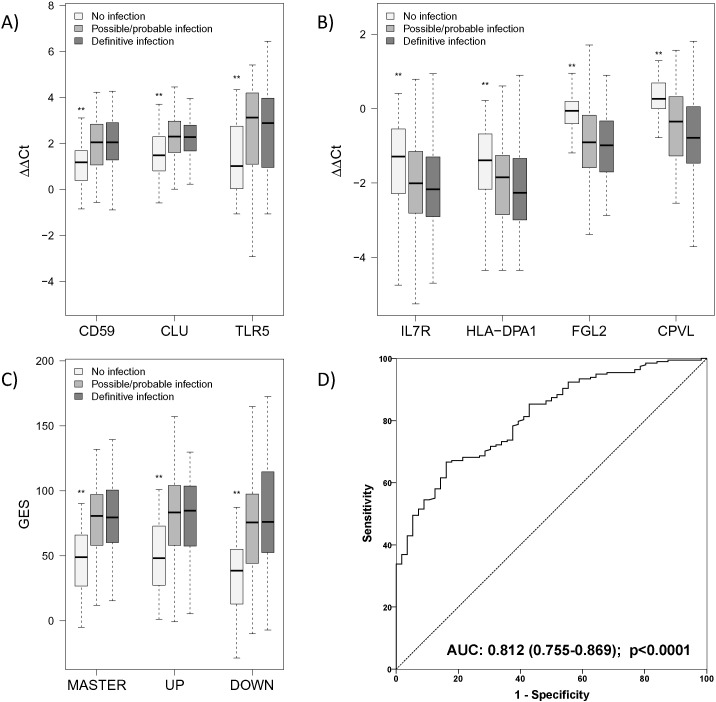
Fig. 4.
Expression characteristics of the individual transcripts of the genomic score and its sub-scores for patients enrolled in the confirmation cohorts. A) Differences to the mean value of a control cohort expressed as ΔΔCt values (Kenneth and Thomas, 2001) (i.e., deviation from group mean of healthy volunteers) for each of the three up-regulated genes forming the UP score (TLR5: Toll-like receptor 5; CD59: Protectin; CLU: Clusterin); B) ΔΔCt values for each of the four down-regulated genes forming the DOWN score (FGL2: Fibrinogen-like 2; HLA-DPA1: Major histocompatibility complex class II, DP alpha1; CPVL: Carboxypeptidase, vitellogenic-like; IL7R: Interleukin-7 receptor); ΔΔCts represent differences of the time to reaction between patients and healthy controls, both normalized to internal reference genes. C) Values of the calculated genomic master score including both aspects of the host response. Biological functions of the individual transcripts are summarized in Table 1. For all markers and scores, significant differences compared with the “no infection” group were confirmed (**p < 0.01), however no significant differences were observed between possible/probable and definitive infection groups (results of the post-hoc pairwise comparison after Kruskal–Wallis test). D) Receiver Operator Characteristics (ROCs) for the genomic master score to differentiate definite and possible/probable infection from no infection.