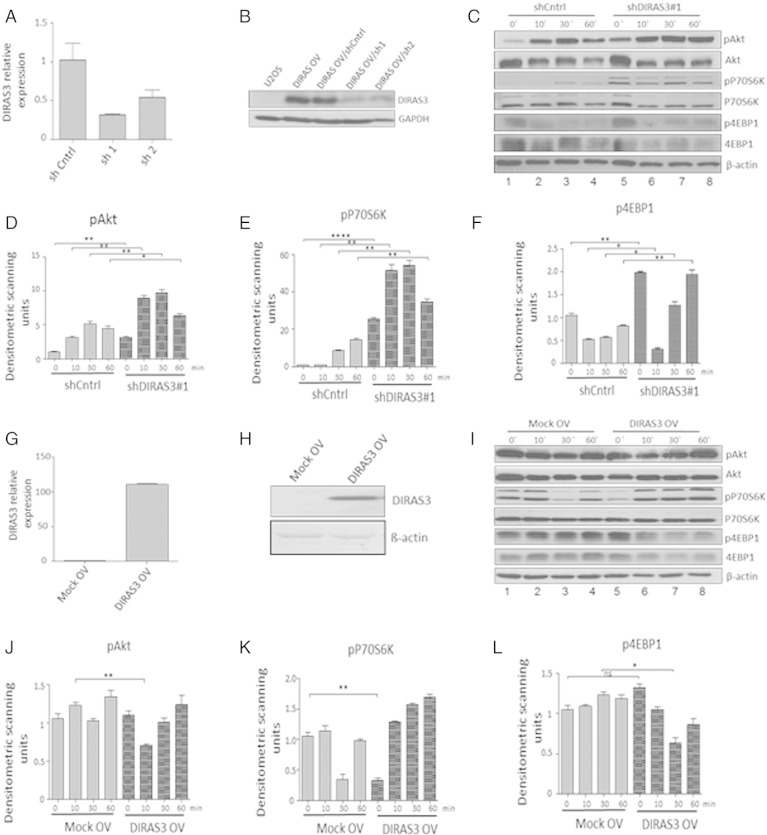
Fig. 2.
Impact of DIRAS3 knock-down (KD) and over-expression (OV) on Akt–mTOR pathway in human ASCs. (A) Efficiency of RNA interference mediated DIRAS3 KD in ASCs was analyzed at mRNA level using q-RT-PCR employing actin as reference gene. (B) DIRAS3 KD efficacy at protein level was evaluated by Western blotting. DIRAS3 was overexpressed in the experimental cell line U-2OS, followed by KD using specific shRNA expressing lentiviruses. (C) DIRAS3 was KD in ASCs using specific shRNA, and cells were density arrested and starved by serum withdrawal for 48 h. Then a cocktail containing 2.5% FCS, insulin, 3-isobutyl-1-methylxanthine, and dexamethasone was added. Cell lysates were harvested at indicated time points. Phosphorylation of Akt (S473), S6K1 (T389) and 4EBP1 (T37/46) was examined by Western blotting. (D–F) Fold changes in densitometric band intensities, acquired by ImageJ were compared. Band intensity of shCntrl at time point 0 min was taken as 1. (G) Overexpression of DIRAS3 mRNA normalized to actin and (H) protein in ASCs infected with lentiviruses expressing DIRAS3 under control of CMV promoter. (I) Following DIRAS3 over-expression, cells were density arrested and starved by serum withdrawal for 48 h. Then a cocktail containing 2.5% FCS, insulin, 3-isobutyl-1-methylxanthine, and dexamethasone was added. Cell lysates were harvested at indicated time points. Phosphorylation of Akt (S473), S6K1 (T389) and 4EBP1 (T37/46) was examined by Western blotting from cell lysates harvested at indicated time points. (J–L) Fold changes in densitometric band intensities, acquired by ImageJ were compared. Band intensity of Mock control at time point 0 min was taken as 1. All error bars represent the means ± SEM. p values * = p < 0.05, ** = p < 0.001 and *** = p < .0001. The significance of difference between means was assessed by analysis of variance (ANOVA) (D, E, F, J, K and L).