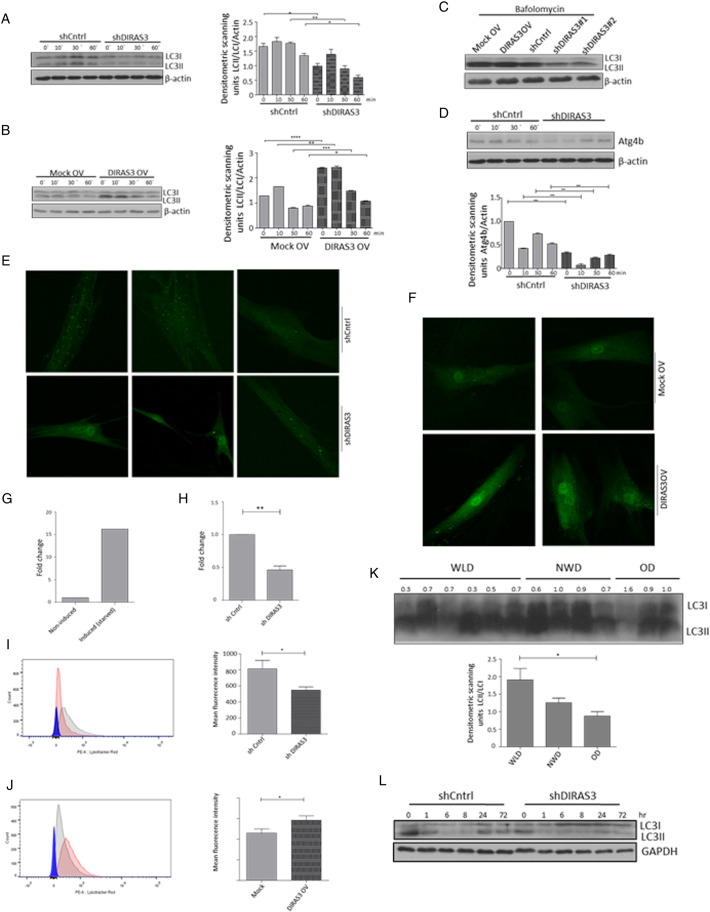
Fig. 5.
WL-induced overexpression of DIRAS3 increases autophagy. (A and B) DIRAS3 was KD (A) or overexpressed (B) in ASCs. (Left panels) Conversion of LC3I to LC3II was monitored by Western blotting at different time points after induction of adipogenesis. (Right panels) Densitometric band intensities were quantified using ImageJ. (C) Autophagy flux was evaluated in DIRAS3 KD and DIRAS3OV ASCs by starving these cells for 3 h with HBSS in the presence of 400 nM bafilomycin A1. Conversion of LC3I to LC3II was monitored by Western blotting. (D) Western blot analysis of ATG4b expression upon DIRAS3 KD (left panel). Band intensities were quantified using ImageJ (right panel). (E and F) ASCs were co-infected with GFP-LC3 and either shDIRAS3 (E) or DIRAS3 overexpressing lentiviruses (F). Cells were starved for 3 h and LC3 punctate formation was visualized by IF-CLSM. 400 × magnification. (G–H) ASCs were co-infected with GFP-LC3 and shControl or shDIRAS3 lentiviruses. Cells were analyzed by image stream multispectral flow cytometer. (G) Serum starved cells were taken as a positive control for autophagy induction and a fold change in autophagy high cells was calculated taking the number of autophagy high cells in the presence of serum as 1. (H) Number of autophagy high cells in shcontrol settings was taken as 1 and a fold change was calculated upon DIRAS3 KD. (I and J) Lysosomal acidity was monitored as a read-out of autophagy in ASCs upon DIRAS3 KD (J) or DIRAS3 OV (K) using Lyso-tracker red dye. (K) WAT lysates from WLDs, NWDs and ODs were analyzed by Western blotting to evaluate LC3I conversion to LC3II (upper panel). Densitometric band intensities were quantified using ImageJ (lower panel). (L) Conversion of LC3I to LC3II was monitored by Western blotting during adipogenesis in DIRAS3 KD and control ASCs. All error bars represent the means ± SEM. p values * = p < 0.05, ** = p < 0.001 and *** = p < .0001. The significance of difference between means was assessed by analysis of variance (ANOVA) (A, B, D and K) and Student's t test (H, I and J).