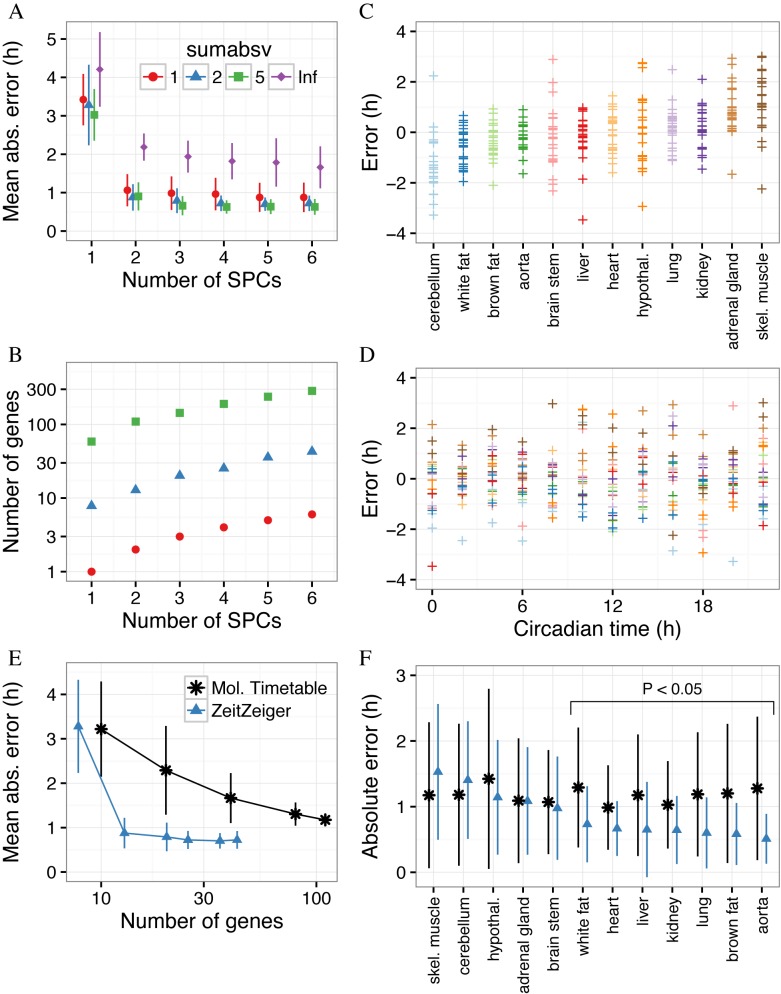
Figure 2.
Using ZeitZeiger to predict circadian time of samples from GSE54650 in leave-one-organ-out cross-validation. (A) Overall mean absolute error (MAE) on cross-validation, as a function of the two main parameters of ZeitZeiger, sumabsv and nSPC. The point shows the overall MAE across all 288 samples and the error bar shows the standard deviation of the MAE across the 12 organs. sumabsv = Inf refers to a predictor trained using standard principal components instead of SPCs. (B) Mean number of genes in the predictors from cross-validation as a function of sumabsv and nSPC. (C) Prediction error (predicted CT minus observed CT) for each organ in GSE54650. Each point is a sample. Organs are sorted by mean error. (D) Prediction error as a function of circadian time. Each point is a sample, with color corresponding to organ, as in C. (E) Mean absolute error versus number of genes for ZeitZeiger and the molecular-timetable method. For ZeitZeiger, the predictor was trained using sumabsv = 2 and various values of nSPC. For the molecular-timetable method, various numbers of genes matching criteria for periodicity and variability were randomly selected. Each point shows the overall MAE across all 288 samples. The error bar shows the standard deviation of the MAE across the 12 organs. Points are connected by straight lines for ease of visualization. (F) Mean absolute error in each organ for ZeitZeiger and the molecular-timetable method. Organs are sorted by decreasing mean absolute error for ZeitZeiger. Statistical significance was evaluated using a two-sided, paired permutation test.