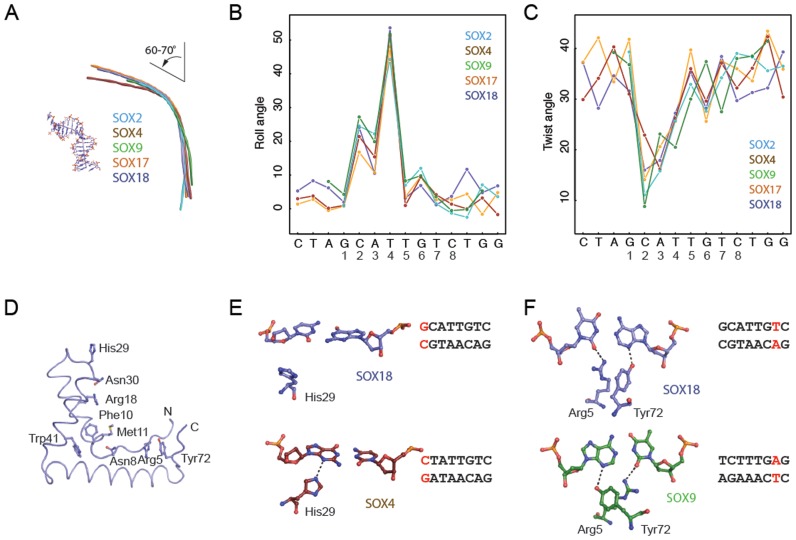
Figure 3.
SOX proteins induce conserved bends and rearrange to accommodate variant DNA elements. (A) Helical axis calculated by Curves+ (43) used to estimate the total bend for SOX2 (PDB ID: 1gt0), SOX4 (3u2b), SOX9 (4euw), SOX17 (3f27) and SOX18 (this study). All five SOX/DNA complexes show a similar overall bend between 60° and 70° (Supplementary Table S4). The inter base-pair parameters roll angle (B) and twist angle (C) were calculated using Curves+ (43) and plotted against the DNA sequence. The x-axis shows the forward strand of the Prox1-DNA sequence co-crystallized with the SOX18-HMG. (D) Tube representation of the SOX18-HMG highlighting the conserved set of DNA-binding residues present in all 20 SOX TFs. DNA was omitted and orientation was changed with respect to Figure 2B for optimized visibility. (E) Comparison of the conformation of His29 in SOX18 (slate blue) and SOX4 (ruby red) and the G1C’1 base-pair (SOX18) which is converted into C1G1’ in the SOX4/DNA complex. (F) Conformational switch of Arg5 and Tyr72 in SOX18 (slate blue) and SOX9 (green) to accommodate the different environment provided by T7A7’ (SOX18) or A7T7’ (SOX9), respectively. H-bonds are indicated with dashed lines. Displayed base pairs are highlighted in red in the core Prox1 elements used.