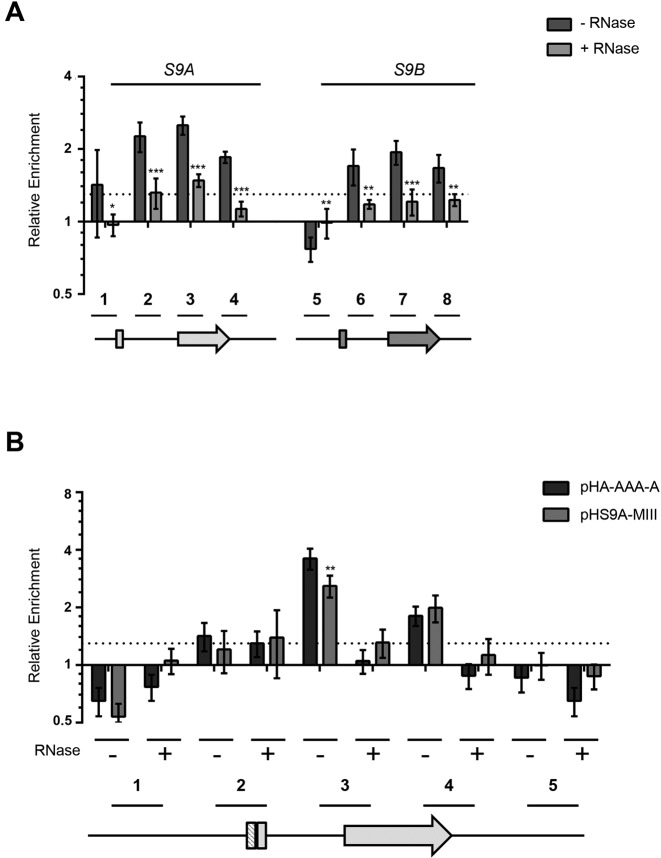
Figure 6.
Rps9B protein binds to the intron of RPS9A in the presence of the two-way helical structure required for inhibiting the splicing of RPS9A pre-mRNA. (A) Rps9B protein differentially interacts with the chromatin of the RPS9 paralogs in an RNA dependent manner. ChIP assays were performed using an HA tagged version of Rps9B isoform before (dark gray bars) or after (light gray bars) RNase treatment and the co-precipitated DNA was detected using paralog specific primers. The enrichment of the different PCR amplicons relative to an untranscribed region in chromosome V is presented in the form of a bar graph. The position of the primers used is shown relative to the paralog gene's structures under the graph. The untranslated regions and introns are shown as lines and exons as boxes. The experiments were conducted in triplicates and the standard deviations are shown as error bars. DNA enrichments were considered significant when greater than 30% relative to the untranscribed control (dotted line). Statistically significant differences between RNase treated and untreated samples are indicated by asterisks (*P-value <0.05, **P-value < 0.01 and ***P-value < 0.001). (B) The Rps9B interaction with RPS9A is enhanced in the presence of the two-way helical structure required for the inhibition of RPS9A expression. The interaction of Rps9B with DNA originating from plasmids carrying wild type RPS9A (pH-A-AAA-A) or RPS9A with mutated two-way helical structure (pHS9A-MIII) was assayed before (–) or after (+) treatment with RNase using ChIP as described in (A). The position of the primers is shown below the graph. Significant differences between the enrichment patterns of the wild type gene and its mutated versions are indicated by asterisks (**P-value < 0.01).