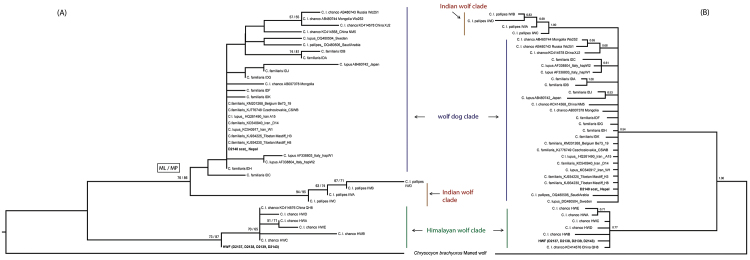
Figure 3.
Phylogenetic trees constructed using 229 bp of aligned CR sequence data. The values at nodes correspond to A bootstrap support > 50% in (ML) and (MP) analyses; and B Bayesian posterior probability > 0.50. Scat samples sequenced in this study are highlighted in bold. Four samples (D2137, D2138, D2139 and D2143) represented a novel haplotype HWF within the Himalayan wolf clade, while a fifth sample (D2140) matched with existing domestic dog haplotypes.