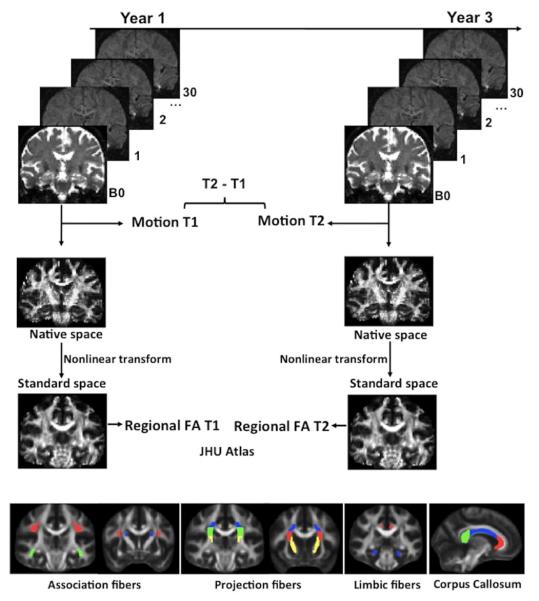
Figure 1. Data processing.
DTI data for each subject were processed independently at each timepoint. DTI scans were pre-processed to generate fractional anisotropy (FA) maps, motion (average rotation and translation parameters) estimates were saved and included in the longitudinal analyses as a time-dependent covariate. FA images were transformed to standard space to compute mean FA in 12 atlas-based regions of interest at each timepoint: Major association fibers (red = superior longitudinal fasciculus, blue = superior frontal occipital fasciculus, green = inferior frontal occipital fasciculus), projection fibers (red = anterior, blue = superior and green = posterior corona radiate, yellow = internal capsule), corpus callosum (red = genu, blue = body and red = splenium) and limbic fibers (red = cingulum and blue = parahippocampal cingulum).