Fig. 4.
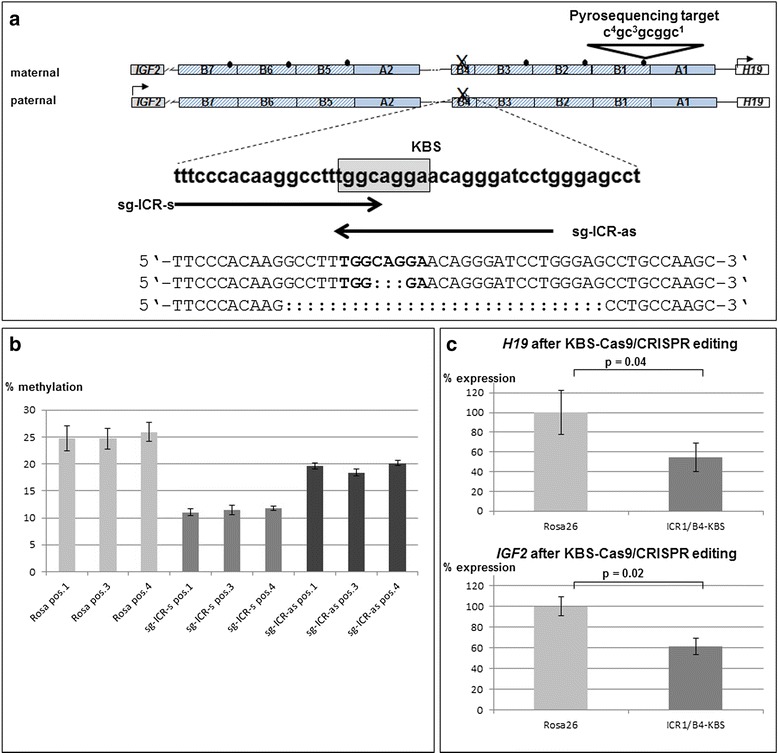
Genomic CRISPR/Cas9 deletion of the ICR1-B4-KBS leads to the loss of ICR1 methylation and transcriptional deregulation of H19 and IGF2. a Sense and antisense sgRNAs (sg-ICR-s, sg-ICR-as) were designed to target the KBS site. Adjacent to both sgRNAs is the presence of a PAM (5′NGG3′). After lentiviral transduction in primary fibroblasts with a differentially methylated ICR1 (50 % methylation), GFP+ cells were sorted by FACS. The target sequence was amplified by PCR and subcloned, and a random selection of 20 clonal integrates was sequenced. Seven out of these 20 clones showed a deletion of the target site. The smallest and largest of the detected Cas9-mediated deletions (ranging from 3 to 29 bp) are depicted here to illustrate the deletion range. All detected deletions were found to disrupt the KBS motif. b Cas9-modified fibroblasts are impaired for ICR1 methylation. For the following analyses, GFP+ cells were sorted by FACS and analysed as pooled cells. Displayed histogram values are represented as mean ± SEM from four replicate analyses for each CG position. c Expression of H19 and IGF2 is altered upon KBS editing. Expression values obtained by qRT-PCR from RNA of pooled FACS-sorted GFP+ cells were normalized against PDH and B2M transcripts and are shown in mean ± SEM from 2 to 3 replicate analyses each
