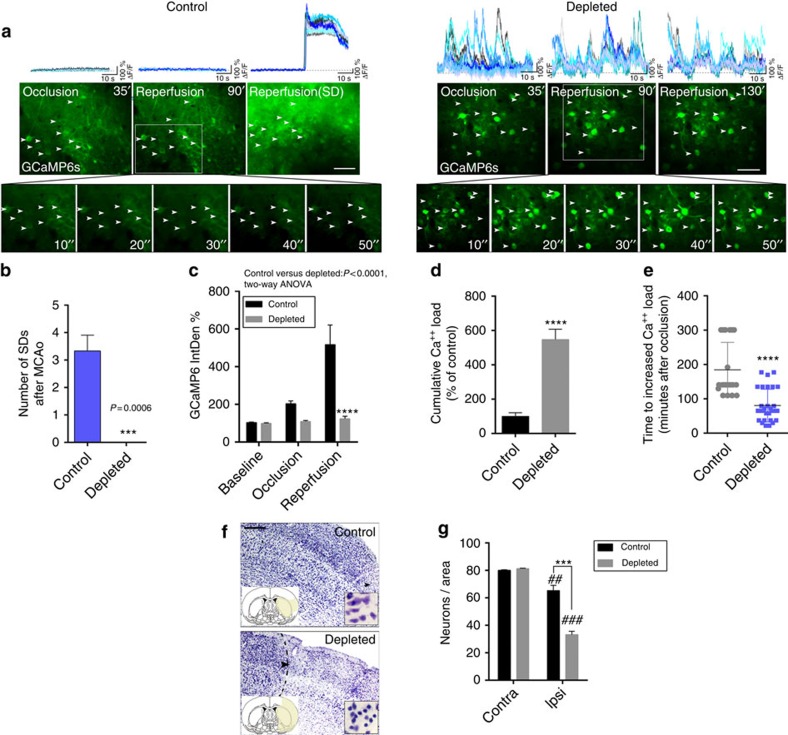
Figure 4. Microglia shape neuronal activity after cerebral ischaemia.
(a) Neuronal GCaMP6s signal changes were assessed by in vivo two-photon imaging in the cerebral cortex using 2 min long recordings with fast resonant scanning (31.25 frames per second), separated by 5 min breaks. In control mice no significant GCaMP6s signal intensity changes were seen before the occurrence of spreading depolarizations (SD) starting typically 1.5–3 h post-reperfusion (left panel, inserts shown at Reperfusion 90′). In contrast, an absence of microglia resulted in slow neuronal oscillations (at ≈0.1 Hz frequency) already during occlusion, with similar changes seen after reperfusion (right panel). Representative calcium transients from 10–10 individual neurons (white arrows, calcium responses are indicated by different colours) are shown on the top panels from control and microglia-depleted mice. (b) Lack of microglia resulted in the absence of SD. (c) Average GCaMP6s intensity of 2 min recordings was increased at late reperfusion in control mice due to initiation of SDs, compared to microglia-depleted animals. Integrated density values were expressed as a percentage of baseline (IntDen%). (d) In contrast, cumulative calcium load (expressed in % of control) as calculated by summing up calcium curve integrales of individual neurons during the 2 min long two-photon recordings over a course of 5.5 h (baseline, 60 min occlusion and 4 h reperfusion) was significantly increased in microglia-depleted animals. (e) Microglia-depleted animals reach increased calcium load over baseline significantly earlier (due to continuous neuronal depolarizations) than control mice (due to delayed initiation of SD). (f) Cresyl violet staining reveals marked neuronal death in the imaging site in microglia-depleted mice compared to controls. (g). Quantification of cresyl violet-stained neurons (N=8 mice, two-way analysis of variance (ANOVA), followed by Sidak's multiple comparison) at the imaging site in the cerebral cortex and the corresponding contralateral hemisphere. Data are expressed as mean±s.e.m. b: N=4 mice per group, unpaired t-test; c: two-way ANOVA followed by Dunn's multiple comparison; d,e: unpaired t-test, c–e: N=52 neurons from 4 individual mice. ##P<0.01, ###P<0.001 versus contralateral, ***P<0.001 control versus depleted. Scale bars, a, 50 μm; f, 200 μm.