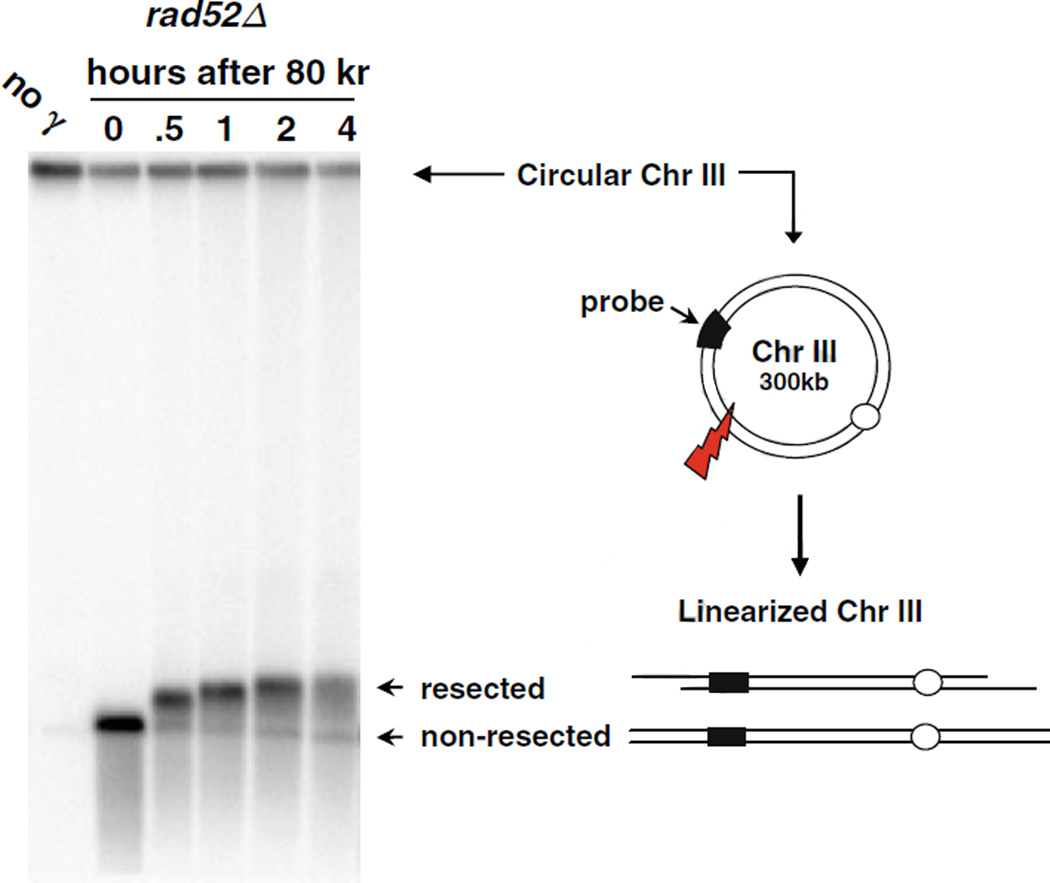
Fig. 2.1.
PFGE-shift of circular chromosomes broken by IR-induced random DSBs. Nocodazole-arrested G2/M rad52Δ cells were irradiated with 80 krads, and post-IR incubation was done in YPDA medium. Cells were collected and prepared at the indicated times. Plug preparation and CHEF parameters are described in text. Chr III was detected by a probe targeting the CHA1 gene. The circular (unbroken) form of Chr III is trapped in the well during PFGE. A single random DSB results in full-length linearized Chr III molecules that can migrate out of the well, forming a unique 300 kb band. The PFGE-shifted DNA corresponding to resected DNA reaches a plateau with “apparent” size of 430 kb. (This image is from 19.)