Figure 1.
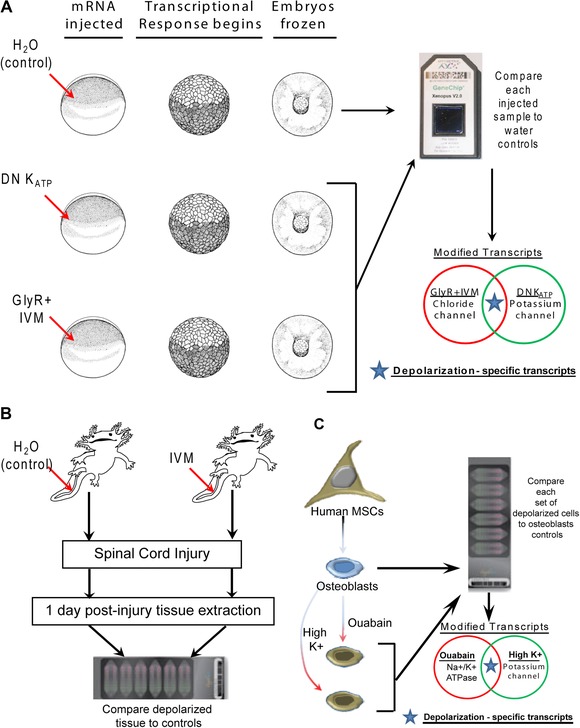
(A) Experimental design for the Xenopus microarray experiment. Xenopus embryos were microinjected at the one‐cell stage with water (control) or mRNA for dominant‐negative KATP (DN KATP, 666 construct) or GlyR channel mRNA. GlyR‐injected embryos were incubated in channel opener drug ivermectin (IVM). The transcriptional response begins in the Xenopus embryos at stage 8. The embryos were flash‐frozen at stage 11, mRNA was extracted, and transcripts were compared between the experimental and control samples. Extracts from 50 embryos was pooled for each experimental group. Only those transcripts that were similarly modified in both the GlyR+IVM and DN KATP groups were used as depolarization‐specific modified transcripts. (B) Experimental design for the axolotl microarray experiment; 2−3 cm axolotl were used. The central canal of the spinal cord was pressure injected with vehicle (water, controls) or injected with IVM (depolarization). Immediately after injection spinal cord injury was performed by removing a small portion of the cord. One day post‐injury the area of injury was removed after anesthetizing the animals. Tissues from 10 animals were pooled for each experiment. Extracted RNA was used to detect changed transcripts in IVM‐injected animals in comparison to control (water injected). (C) Experimental design for the primary human mesenchymal stem cells (hMSCs) microarray experiment. hMSCs were induced to differentiate into osteoblasts. These osteoblasts were then treated with ouabain (Na+/K+ ATPase inhibitor) or incubated in medium with high potassium (both depolarizing conditions). The mRNA extracted from the treated cells was compared with that of untreated osteoblasts (controls). Among the modified transcripts only those that were similarly modified in both the treatments were used as depolarization‐specific modified transcripts.
