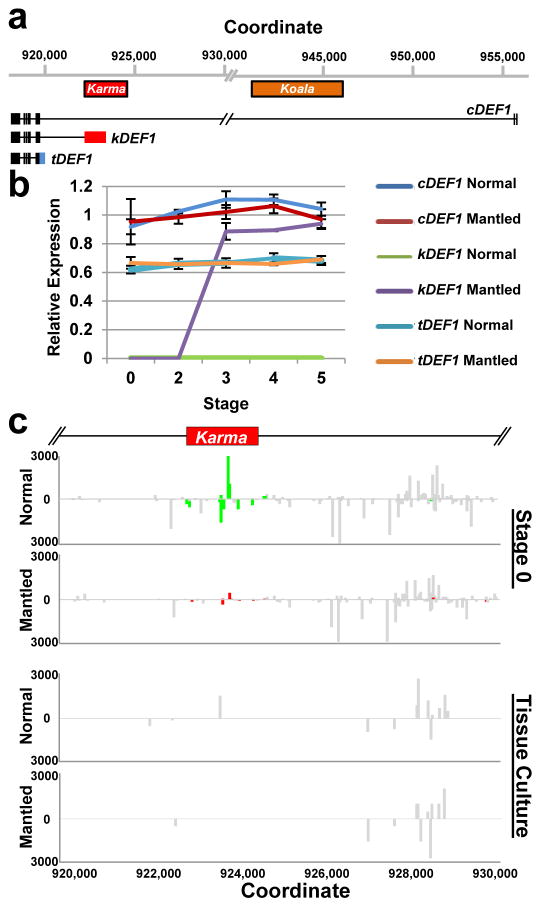
Figure 4. Alternative splicing and loss of 24nt small RNA.
a, EgDEF1/MANTLED transcripts assembled from transcriptome sequencing (data not shown) and RT-PCR (Methods). Black boxes, exons. Blue box, intron 5 sequence included in the tDEF1 transcript. Coordinates relative to the reference pisifera oil palm genome14. b, Quantitative RT-PCR of cDEF1, tDEF1 and kDEF1 transcripts in shoot apices (stage 0) and in early (stage 2) to late (stage 5) female inflorescences from normal and parthenocarpic mantled ramets. Error bars, standard deviations between 3 replicate assays of 3 replicate tissue samples per phenotype, per stage. Expression relative to an endogenous reference gene is shown (Methods). c, 24nt siRNA accumulation in shoot apices (stage 0) from normal (n=5) and parthenocarpic mantled (n=7) ramets, and from second passage apical leaf tissue cultures re-cloned from normal (n=2) or mantled (n=1) ramets (Methods). Values expressed as fragments per kilobase per million mapped reads (FPKM). Bars above (sense) and below (antisense) the line indicate mapped normalized 24nt siRNAs that are not significantly different in abundance in normal and mantled (grey) or significantly differentially expressed in normal (green) relative to mantled (red) (p <0.05, Student’s t-test, two tailed, assuming equal variance).