Fig 2. Phylogenetic tree highlighting the position of Borrelia recurrentis identified in the present study relative to Borrelia type strains and uncultured borreliae.
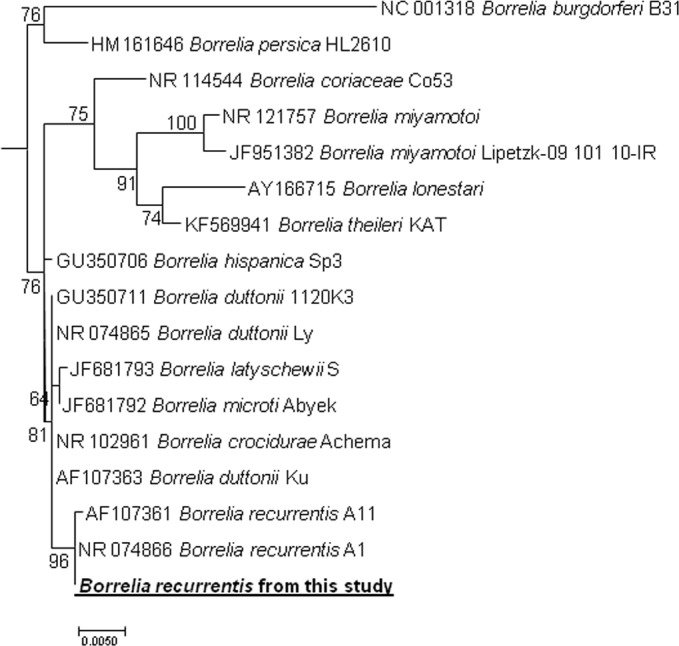
The 16S rRNA gene sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained from a maximum likelihood (ML) phylogenetic analysis with the TrN+Γ substitution model. The GenBank accession numbers are indicated at the beginning. Sequence obtained in the present study is in bold. The numbers at the nodes are the bootstrap values obtained by repeating the analysis 100 times to generate a majority consensus tree. There were a total of 1,269 positions in the final dataset. The scale bar indicates a 5% nucleotide sequence divergence.
