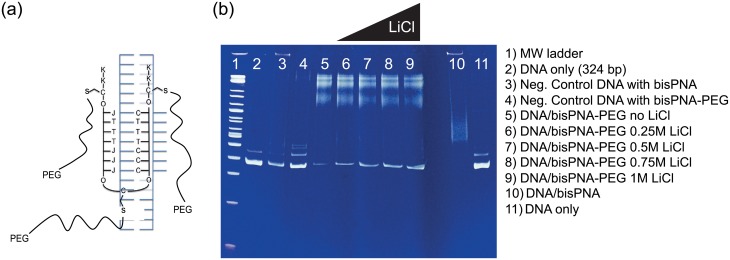
Fig 2. The bisPNA probe binds to dsDNA in conditions compatible with nanopore experiments, with and without a PEG payload.
(a) The cysteine substituted bisPNA (black, U-shape) is bound to 324 bp scaffold dsDNA (blue) making a triplex helix. A modified cytosine is used that has less pH dependence when making Hoogsteen contacts (J). The two halves of the PNA are separated by a flexible PEG linker (O) that has a cysteine (C) amino acid in the middle. Lysines (K) are added at each end to increase the stability. PEGs containing maleimides react with the cysteine residues (C) in the PNA creating the DNA/bisPNA-PEG complex. (b) The DNA/bisPNA complex was found to be stable in up to 1M LiCl over a half-hour incubation at room temperature, indicating that the majority of the complex will be intact throughout the duration of the nanopore assay. Image shows 10% PAGE EMSA with lanes: 1) high molecular weight ladder, 2) DNA (324 bp), 3) DNA (negative control with scrambled 7 bp target sequence) with PNA, 4) DNA (negative control) with PNA-PEG (5 kDa), 5) DNA/PNA-PEG (5 kDa) and 10) DNA/PNA. Lanes (6-9) are DNA/PNA-PEG (5 kDa) after 30 min incubation in increasing LiCl concentrations (0.25, 0.5, 0.75, 1M).