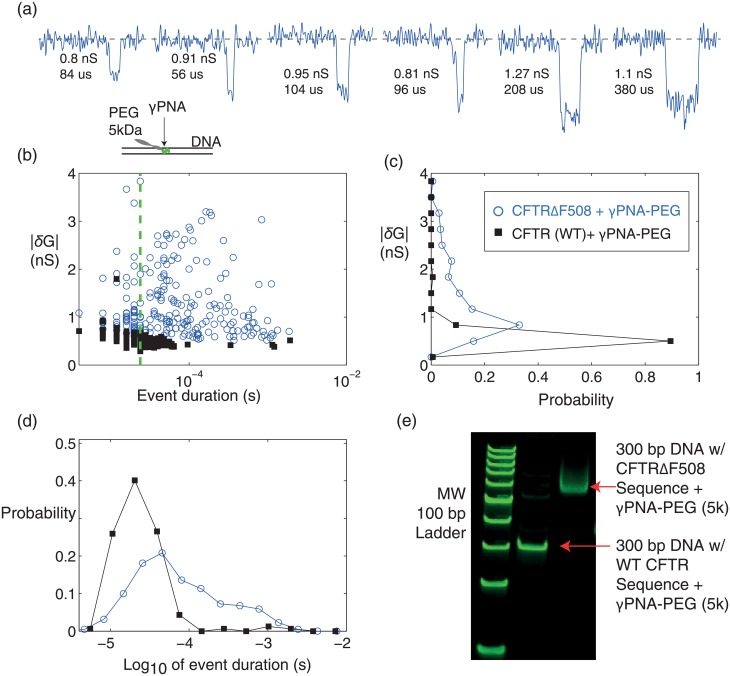
Fig 6. The 300 bp DNA/γPNA-PEG 5 kDa complex is resolvable with a 26-36 nm diameter pore in 100 mM LiCl, providing positive detection of the CFTRΔF508 gene mutation.
(a) Representative events with the pore initially at 26 nm in diameter, reporting δG and duration values. (b) Population of δG vs. duration for all 221 events over 52 minutes at 1 nM complex and 200 mV. Events span three pore sizes (26 nm, 32nm, 36 nm) that were enlarged by dielectric breakdown. The green line (24 μsec) is the minimum duration for resolving δG. (c) δG histogram and (d) duration histogram of all events. (e) 5% PAGE EMSA shows the 22 bp γPNA-PEG (5k) bound to the 300 bp DNA at the 22 bp target sequence that encompasses the CFTRΔF508 mutation (right lane). The γPNA-PEG (5k) does not bind to the 300 bp DNA that has the wild-type (i.e., non-mutant) sequence (middle lane), showing target specificity. The sizing ladder is low molecular weight (left lane).