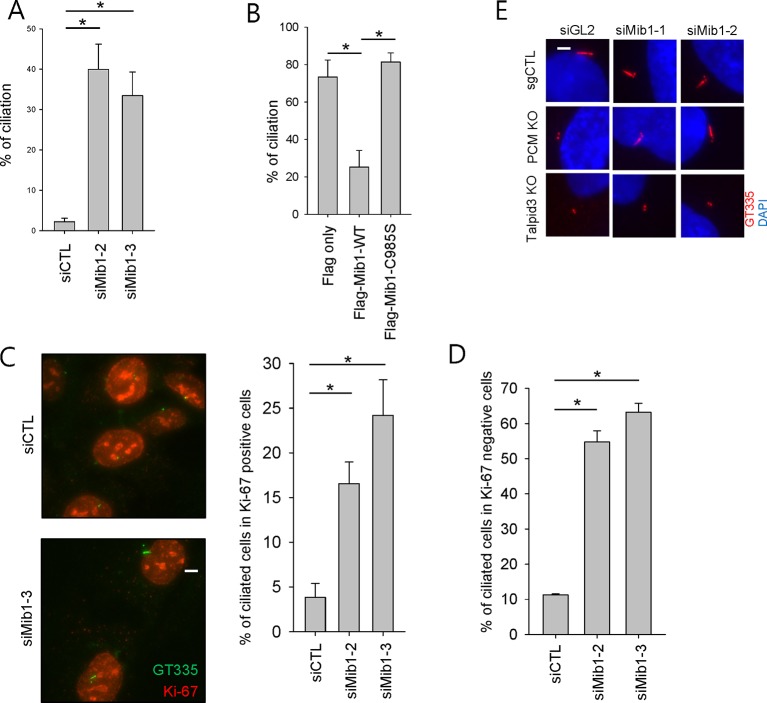
Figure 5. Mib1 is downstream of PCM1.
(A) Validation of siRNA knockdown of Mib1. Control (sgCTL), PCM1, and TALPID3 KO RPE1 cells were transfected with the indicated siRNAs corresponding to non-specific control (siGL2) or Mib1 (siMIB1-1 or siMIB1-2), and lysates were subjected to western blot analysis with the indicated antibodies. (B) Mib1 depletion partially rescues the ciliogenesis defect in PCM1 KO cells. Cell lines in panel A were transfected with siRNAs corresponding to non-specific control (siGL2) or Mib1 (siMIB1-1 or siMIB1-2) for 48 hr and serum starved for 48 hr. Ciliated cells (n ≥ 100 per sample in three independent experiments) were analyzed by immuno-staining with GT335. Error bars, SEM. *p<0.05. (C) E3 ligase activity of Mib1 underlies ciliogenesis defect in PCM1 KO cells. Control and PCM1 KO RPE1 cells were infected with empty (EV), Flag-Mib1-WT or Flag-Mib1-C985S lentiviruses for 72 hr and serum starved for 48 hr. Ciliated cells were analyzed by immuno-staining with GT335; n≥100 per sample were analyzed in two independent experiments. Error bars, SD. *p<0.05. (D) Control, PCM1, and Talpid3 KO RPE1 cells were transfected with siGL2, siMIB1-2, or siMIB1-3 for 48 hr and subjected to western blot analysis with the indicated antibodies.