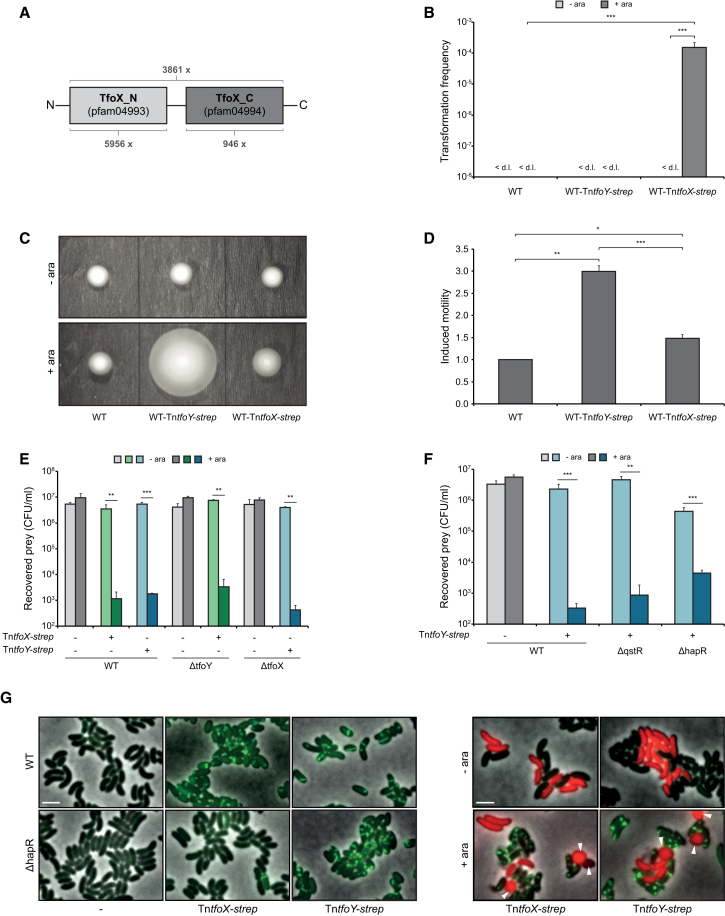
Figure 1.
TfoX and TfoY Are Not Redundant
(A) Scheme showing the two-domain structure of TfoX-like proteins. The abundance of proteins carrying either one or both domains is indicated below and above the scheme, respectively.
(B–D) V. cholerae strains carrying a chromosomal copy of either tfoX or tfoY under the control of PBAD (TntfoX-strep and TntfoY-strep) were analyzed for chitin-independent natural transformability (B) and motility on semi-solid LB agar (C and D). (B) Natural transformation is fully dependent on TfoX. The indicated strains were cultured under non-inducing (−ara) and inducing (+ara) conditions. Transformation frequencies are shown on the y axis and represent the average of three independent experiments (error bars indicate SD). <d.l., below detection limit. (C and D) TfoY induction vastly enhances surface motility. Motility was scored on soft agar without (−ara) and with (+ara) induction. Representative images are shown (C). (D) Quantification of the motility phenotype shown in (C). The average ratio between induced versus uninduced conditions is shown on the y axis based on three independent experiments (±SD).
(E–G) TfoY induces T6SS-mediated interbacterial killing in a TfoX- and QS-independent manner. (E and F) Interspecies killing assay between V. cholerae strains and E. coli as prey. Indicated V. cholerae were co-cultured with the prey on plain LB agar (−ara) or LB agar plates supplemented with arabinose (+ara) to induce tfoX (green) or tfoY (blue). The survival of the prey is depicted as colony-forming units (CFU) per ml. Data represent the average of at least three independent biological replicates (±SD). (G) Visualization of T6SS structures (left) and T6SS-induced cell rounding of prey (right) by fluorescence microscopy. Attacked rounded prey cells are indicated by arrowheads. The brightness of the stronger TfoX-induced VipA-sfGFP signal was reduced for better visualization. Statistical significance is indicated for all panels (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
See also Figure S1.