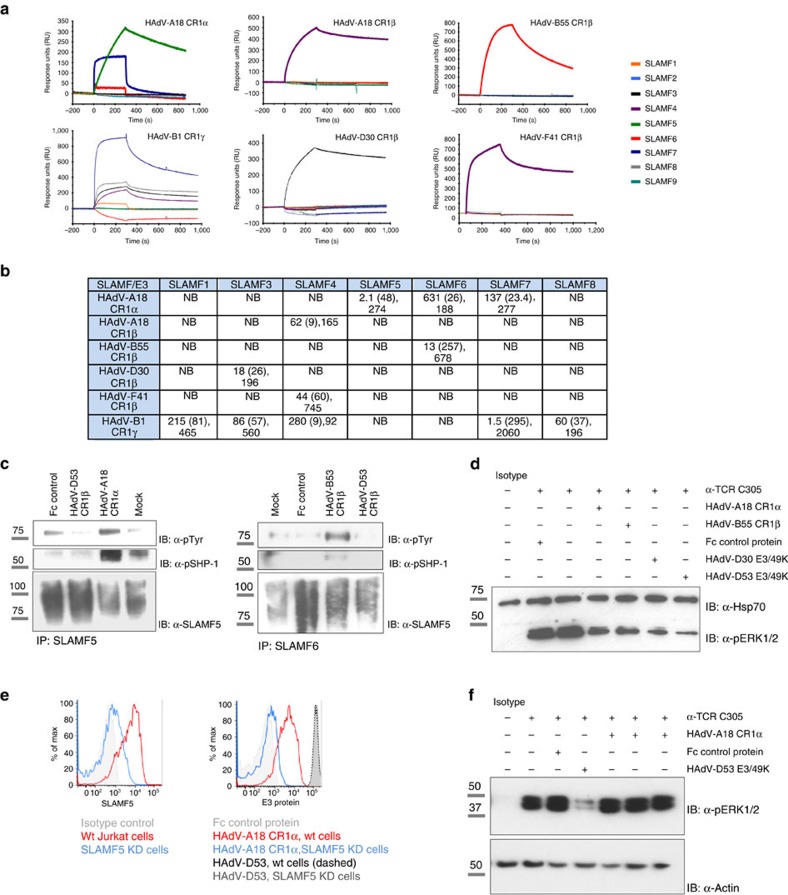
Figure 4. SLAM receptor family is a hub for E3-mediated interference.
(a) Analysis of SLAM receptor ECDs (400 nM) binding to the E3 proteins indicated, immobilized on SPR chips. (b) Kinetic parameters calculated for the interaction between the E3 proteins and the SLAM receptors targeted. Binding affinities are shown as KD (nM) values. The χ2, in parenthesis, followed by and Rmax values indicate the goodness of the experimental fitting. NB, no binding. (c) HEL cells were stimulated as indicated and cell lysates were immunoprecipitated for SLAMF5 (left) or SLAMF6 (right), and immunoblotted to detect phosphorylation of tyrosine residues, SLAMF5, SLAMF6 or phosphoSHP-1. An irrelevant protein carrying the same tag (human Fc) as the viral proteins, named ‘Fc control’, was included in all the experiments throughout the manuscript to control for potential nonspecific effects due to the presence of this tag. (d) Jurkat cells were pre-incubated with control or E3 proteins before stimulation with an anti-TCR antibody and phosphorylation of ERK1/2 was analysed by immunoblotting. (e) Expression of SLAMF5 on the surface of wt or SLAMF5 knockdown (KD) Jurkat cells (left histogram) and binding of the indicated viral proteins to both cell types (right histogram). HAdV-D53 E3/49K binds to CD45 and was used as a control. (f) SLAMF5 KD cells were stimulated as in d to analyse the implication of this receptor in HAdV-A18 CR1α-mediated inhibition. The HAdV-A18 CR1α protein was assayed at 1, 10 and 50 nM (from left to right), whereas control proteins were added at 50 nM. Assays are representative of at least three independent experiments. Molecular masses are indicated in kDa. Full immunoblots are shown in Supplementary Fig. 8b–e.