Figure 1.
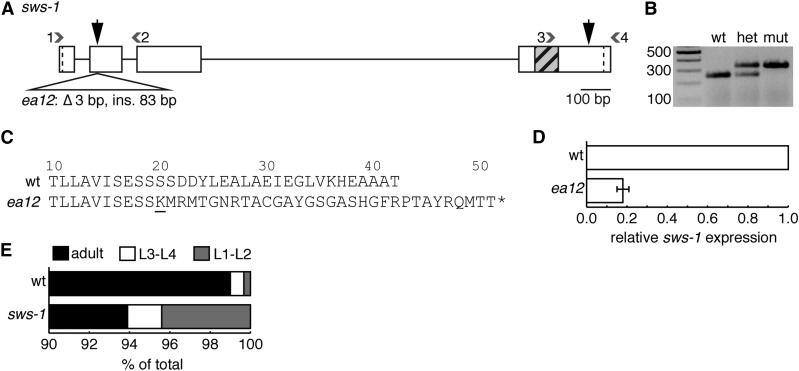
sws-1(ea12) is an insertion/deletion that results in an early stop codon. (A) Diagram of sws-1-coding region. Boxes and straight lines represent exons and introns, respectively. Start and stop codons are demarcated by dashed lines. Gray hatched box shows DNA encoding the SWIM domain. Large black vertical arrows mark predicted Cas9 cleavage sites for each injected gRNA; small gray numbered arrowheads represent primers used for screening (primer sequences are listed in Table S2). ea12 is a 3-bp deletion/83-bp insertion in exon 2. (B) Representative image of ea12 genotyping using primer combination 1 and 2 as shown in A. The mutant allele is readily detected as the slower migrating band on a 2% agarose gel. (C) Predicted protein sequence of exon 2 of wt (top) and ea12 (bottom) SWS-1. sws-1(ea12) is predicted to produce the first 19 amino acids of the wild-type SWS-1 protein followed by 32 frameshifted amino acids prior to truncation (underlined letter marks beginning of frameshift). (D) Expression of sws-1 mRNA in wild-type and sws-1(ea12) hermaphrodites. The data are presented as the mean expression of sws-1 relative to reference gene rpl-32 ± SEM for two biological replicates. (E) Developmental progression of wild type and sws-1. For each genotype, 100 L1’s were plated in triplicate and scored 50 hr later as L1–L2, L3–L4, or adult. The results shown are the percentage of total worms in each developmental stage. A subset of sws-1 mutants arrested as L1–L2 larvae (P < 0.001 vs. wt, Fisher’s exact test).